Mangrove soil as a source for novel xylanase and amylase as determined by cultivation-dependent and cultivation-independent methods
- PMID: 31741310
- PMCID: PMC7058812
- DOI: 10.1007/s42770-019-00162-7
Mangrove soil as a source for novel xylanase and amylase as determined by cultivation-dependent and cultivation-independent methods
Abstract
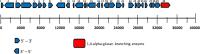
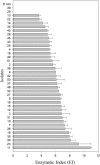
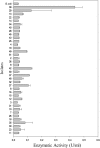
Xylanase and α-amylase enzymes participate in the degradation of organic matter, acting in hemicellulose and starch mineralization, respectively, and are in high demand for industrial use. Mangroves represent a promising source for bioprospecting enzymes due to their unique characteristics, such as fluctuations in oxic/anoxic conditions and salinity. In this context, the present work aimed to bioprospect xylanases from mangrove soil using cultivation-dependent and cultivation-independent methods. Through screening from a metagenomic library, three potentially xylanolytic clones were obtained and sequenced, and reads were assembled into contigs and annotated. The contig MgrBr135 was affiliated with the Planctomycetaceae family and was one of 30 ORFs selected for subcloning that demonstrated only amylase activity. Through the cultivation method, 38 bacterial isolates with xylanolytic activity were isolated. Isolate 11 showed an enzymatic index of 10.9 using the plate assay method. Isolate 39 achieved an enzyme activity of 0.43 U/mL using the colorimetric method with 3,5-dinitrosalicylic acid. Isolate 39 produced xylanase on culture medium with salinity ranging from 1.25 to 5%. Partial 16S rRNA gene sequencing identified isolates in the Bacillus and Paenibacillus genera. The results of this study highlight the importance of mangroves as an enzyme source and show that bacterial groups can be used for starch and hemicellulose degradation.
Keywords: Biotechnology; Culture-dependent method; Enzyme; Metagenomic library.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

Similar articles
-
Functional metagenomics of oil-impacted mangrove sediments reveals high abundance of hydrolases of biotechnological interest.World J Microbiol Biotechnol. 2017 Jul;33(7):141. doi: 10.1007/s11274-017-2307-5. Epub 2017 Jun 7. World J Microbiol Biotechnol. 2017. PMID: 28593475
-
Deciphering the role of Paenibacillus strain Q8 in the organic matter recycling in the acid mine drainage of Carnoulès.Microb Cell Fact. 2012 Feb 3;11:16. doi: 10.1186/1475-2859-11-16. Microb Cell Fact. 2012. PMID: 22305268 Free PMC article.
-
Identification and functional characterization of an α-amylase with broad temperature and pH stability from Paenibacillus sp.Appl Biochem Biotechnol. 2013 May;170(2):359-69. doi: 10.1007/s12010-013-0197-z. Epub 2013 Mar 26. Appl Biochem Biotechnol. 2013. PMID: 23526111
-
Alkalistable xylanase production by alkalitolerant Paenibacillus montaniterrae RMV1 isolated from red mud.J Basic Microbiol. 2014 Oct;54(10):1023-9. doi: 10.1002/jobm.201300357. Epub 2013 Sep 11. J Basic Microbiol. 2014. PMID: 24026904
-
Planctomycetes in boreal and subarctic wetlands: diversity patterns and potential ecological functions.FEMS Microbiol Ecol. 2019 Feb 1;95(2). doi: 10.1093/femsec/fiy227. FEMS Microbiol Ecol. 2019. PMID: 30476049 Review.
Cited by
-
Xylanolytic Extremozymes Retrieved From Environmental Metagenomes: Characteristics, Genetic Engineering, and Applications.Front Microbiol. 2020 Sep 17;11:551109. doi: 10.3389/fmicb.2020.551109. eCollection 2020. Front Microbiol. 2020. PMID: 33042057 Free PMC article. Review.
-
Characterization of a novel GH10 alkali-thermostable xylanase from a termite microbiome.Bioresour Bioprocess. 2022 Aug 17;9(1):84. doi: 10.1186/s40643-022-00572-w. Bioresour Bioprocess. 2022. PMID: 38647897 Free PMC article.
-
Genome sequence of an uncharted halophilic bacterium Robertkochia marina with deciphering its phosphate-solubilizing ability.Braz J Microbiol. 2021 Mar;52(1):251-256. doi: 10.1007/s42770-020-00401-2. Epub 2020 Nov 3. Braz J Microbiol. 2021. PMID: 33141351 Free PMC article.
-
Lignocellulolytic Biocatalysts: The Main Players Involved in Multiple Biotechnological Processes for Biomass Valorization.Microorganisms. 2023 Jan 8;11(1):162. doi: 10.3390/microorganisms11010162. Microorganisms. 2023. PMID: 36677454 Free PMC article. Review.
References
-
- Schaeffer-Novelli Y, Cintrdn-Molero G, Soares MLG, De-Rosa T. Brazilian mangroves. Aquat Ecosyst Health Manag. 2000;3:561–570. doi: 10.1080/14634980008650693. - DOI
-
- Thatoi H, Behera BC, Mishra RR, Dutta SK. Biodiversity and biotechnological potential of microorganisms from mangrove ecosystems: a review. Ann Microbiol. 2013;63:1–19. doi: 10.1007/s13213-012-0442-7. - DOI
-
- Sermsathanaswadi J, Baramee S, Tachaapaikoon C, et al. The family 22 carbohydrate-binding module of bifunctional xylanase/β-glucanase Xyn10E from Paenibacillus curdlanolyticus B-6 has an important role in lignocellulose degradation. Enzym Microb Technol. 2017;96:75–84. doi: 10.1016/j.enzmictec.2016.09.015. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

