Inactivation of cardiotrophin-like cytokine, a second ligand for ciliary neurotrophic factor receptor, leads to cold-induced sweating syndrome in a patient
- PMID: 16782820
- PMCID: PMC1502507
- DOI: 10.1073/pnas.0509598103
Inactivation of cardiotrophin-like cytokine, a second ligand for ciliary neurotrophic factor receptor, leads to cold-induced sweating syndrome in a patient
Abstract
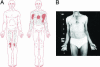
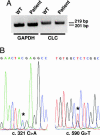
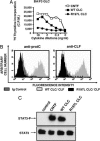
Ciliary neurotrophic factor (CNTF) receptor controls a pathway supporting the differentiation and survival of a wide range of neural cell types during development and in adulthood. Cardiotrophin-like cytokine (CLC)-cytokine-like factor 1 (CLF) composite cytokine is a second ligand for the CNTF alpha-component receptor (CNTFRalpha). This composite cytokine is built on the structural model of IL-12, with a complex formed by a four-helix bundle type I cytokine, CLC (also referred to as CLCF1), bound to a soluble receptor subunit, CLF (also known as CRLF1). We have reported mutations in the chaperone soluble receptor CLF, causing cold-induced sweating syndrome (CISS). In this study, we studied the CLC-mutated alleles in a patient suffering from a similar disease. This patient was compound heterozygous for two different CLC mutations. The first allele was inactivated by a stop codon at position 107 (Y107X). In the second allele, a R197L mutation in the CLC-predicted binding site to the CNTFRalpha was detected. Functional analysis of the mutated protein revealed an incapacity for R197L CLC to bind to CNTFRalpha and activate the subsequent signaling events. Structural and docking interaction studies showed that the R197L substitution destabilized the contact site between CLC and CNTFRalpha.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures

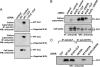
Similar articles
-
The cytokines cardiotrophin-like cytokine/cytokine-like factor-1 (CLC/CLF) and ciliary neurotrophic factor (CNTF) differ in their receptor specificities.Cytokine. 2012 Dec;60(3):653-60. doi: 10.1016/j.cyto.2012.08.014. Epub 2012 Sep 15. Cytokine. 2012. PMID: 22986012
-
The ciliary neurotrophic factor receptor alpha component induces the secretion of and is required for functional responses to cardiotrophin-like cytokine.EMBO J. 2001 Apr 2;20(7):1692-703. doi: 10.1093/emboj/20.7.1692. EMBO J. 2001. PMID: 11285233 Free PMC article.
-
Cytokine-Like Factor 1, an Essential Facilitator of Cardiotrophin-Like Cytokine:Ciliary Neurotrophic Factor Receptor α Signaling and sorLA-Mediated Turnover.Mol Cell Biol. 2016 Mar 31;36(8):1272-86. doi: 10.1128/MCB.00917-15. Print 2016 Apr. Mol Cell Biol. 2016. PMID: 26858303 Free PMC article.
-
CRLF1 and CLCF1 in Development, Health and Disease.Int J Mol Sci. 2022 Jan 17;23(2):992. doi: 10.3390/ijms23020992. Int J Mol Sci. 2022. PMID: 35055176 Free PMC article. Review.
-
The cytokine receptor gp130: faithfully promiscuous.Sci STKE. 2003 Sep 23;2003(201):PE40. doi: 10.1126/stke.2003.201.pe40. Sci STKE. 2003. PMID: 14506288 Review.
Cited by
-
Differential secretion of the mutated protein is a major component affecting phenotypic severity in CRLF1-associated disorders.Eur J Hum Genet. 2011 May;19(5):525-33. doi: 10.1038/ejhg.2010.253. Epub 2011 Feb 16. Eur J Hum Genet. 2011. PMID: 21326283 Free PMC article.
-
Janus kinase 2/signal transducer and activator of transcription 3 inhibitors attenuate the effect of cardiotrophin-like cytokine factor 1 and human focal segmental glomerulosclerosis serum on glomerular filtration barrier.Transl Res. 2015 Oct;166(4):384-98. doi: 10.1016/j.trsl.2015.03.002. Epub 2015 Mar 16. Transl Res. 2015. PMID: 25843671 Free PMC article.
-
Induction of neuro-protective/regenerative genes in stem cells infiltrating post-ischemic brain tissue.Exp Transl Stroke Med. 2010 May 28;2(1):11. doi: 10.1186/2040-7378-2-11. Exp Transl Stroke Med. 2010. PMID: 20509949 Free PMC article.
-
Renal and Hematological Effects of CLCF-1, a B-Cell-Stimulating Cytokine of the IL-6 Family.J Immunol Res. 2015;2015:714964. doi: 10.1155/2015/714964. Epub 2015 Jun 4. J Immunol Res. 2015. PMID: 26146641 Free PMC article.
-
Cardiotrophin-like cytokine (CLCF1) modulates mesenchymal stem cell osteoblastic differentiation.J Biol Chem. 2019 Aug 9;294(32):11952-11959. doi: 10.1074/jbc.AC119.008361. Epub 2019 Jun 27. J Biol Chem. 2019. PMID: 31248987 Free PMC article.
References
-
- Ip N. Y., Yancopoulos G. D. Annu. Rev. Neurosci. 1996;19:491–515. - PubMed
-
- Ip N. Y., McClain J., Barrezueta N. X., Aldrich T. H., Pan L., Li Y., Wiegand S. J., Friedman B., Davis S., Yancopoulos G. D. Neuron. 1993;10:89–102. - PubMed
-
- Sendtner M., Schmalbruch H., Stockli K. A., Carroll P., Kreutzberg G. W., Thoenen H. Nature. 1992;358:502–504. - PubMed
-
- Mitsumoto H., Ikeda K., Klinkosz B., Cedarbaum J. M., Wong V., Lindsay R. M. Science. 1994;265:1107–1110. - PubMed
-
- Helgren M. E., Squinto S. P., Davis H. L., Parry D. J., Boulton T. G., Heck C. S., Zhu Y., Yancopoulos G. D., Lindsay R. M., DiStefano P. S. Cell. 1994;76:493–504. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

