Conventional and microfluidic methods: Design and optimization of lipid-polymeric hybrid nanoparticles for gene therapy
- PMID: 38872047
- PMCID: PMC11782348
- DOI: 10.1007/s13346-024-01644-4
Conventional and microfluidic methods: Design and optimization of lipid-polymeric hybrid nanoparticles for gene therapy
Abstract
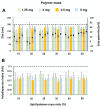
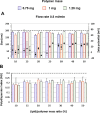
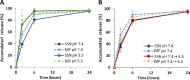
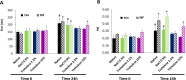
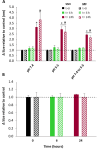
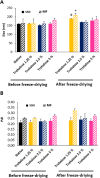
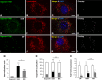
Gene therapy holds significant promise as a therapeutic approach for addressing a diverse range of diseases through the suppression of overexpressed proteins and the restoration of impaired cell functions. Developing a nanocarrier that can efficiently load and release genetic material into cells remains a challenge. The primary goal of this study is to develop formulations aimed to enhance the therapeutic potential of GapmeRs through technological approaches. To this end, lipid-polymeric hybrid nanoparticles (LPHNPs) with PLGA, DC-cholesterol, and DOPE-mPEG2000 were produced by conventional single-step nanoprecipitation (SSN) and microfluidic (MF) methods. The optimized nanoparticles by SSN have a size of 149.9 ± 18.07 nm, a polydispersity index (PdI) of 0.23 ± 0.02, and a zeta potential of (ZP) of 29.34 ± 2.44 mV, while by MF the size was 179.8 ± 6.3, a PdI of 0.24 ± 0.01, and a ZP of 32.25 ± 1.36 mV. Furthermore, LPHNPs prepared with GapmeR-protamine by both methods exhibit a high encapsulation efficiency of approximately 90%. The encapsulated GapmeR is completely released in 24 h. The LPHNP suspensions are stable for up to 6 h in 10% FBS at pH 5.4 and 7.4. By contrast, LPHNPs remain stable in suspension in 4.5% albumin at pH 7.4 for 24 h. Additionally, LPHNPs were successfully freeze-dried using trehalose in the range of 2.5-5% as cryoprotectant The LPHNPs produced by MF and SSN increase, 6 and 12 fold respectively, GapmeR cell uptake, and both of them reduce by 60-70% expression of Tob1 in 48 h.Our study demonstrates the efficacy of the developed LPHNPs as carriers for oligonucleotide delivery, offering valuable insights for their scale up production from a conventional bulk methodology to a high-throughput microfluidic technology.
Keywords: Endosomal escape; Gene therapy; Lipid-polymeric hybrid nanoparticles; Microfluidics; Nanoparticles stability.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval: Not applicable. Competing interests: The authors have non-financial interests to disclose.
Figures


Similar articles
-
pH sensitive lipid polymeric hybrid nanoparticle (LPHNP) of paclitaxel and curcumin for targeted delivery in breast cancer.Drug Dev Ind Pharm. 2024 Oct;50(10):856-864. doi: 10.1080/03639045.2024.2421198. Epub 2024 Nov 3. Drug Dev Ind Pharm. 2024. PMID: 39461888
-
Peer Play.2023 Jul 4. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2023 Jul 4. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30020595 Free Books & Documents.
-
Exploring conceptual and theoretical frameworks for nurse practitioner education: a scoping review protocol.JBI Database System Rev Implement Rep. 2015 Oct;13(10):146-55. doi: 10.11124/jbisrir-2015-2150. JBI Database System Rev Implement Rep. 2015. PMID: 26571290
-
Synthesis and Characterization of Thallium-Texaphyrin Nanoparticles and Their Assessment as Potential Delivery Systems for Auger Electron-Emitting 201Tl to Cancer Cells.Mol Pharm. 2025 Jan 6;22(1):242-254. doi: 10.1021/acs.molpharmaceut.4c00873. Epub 2024 Dec 16. Mol Pharm. 2025. PMID: 39681352 Free PMC article.
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
References
-
- Merz L, Hobel S, Kallendrusch S, Ewe A, Bechmann I, Franke H, et al. Tumor tissue slice cultures as a platform for analyzing tissue-penetration and biological activities of nanoparticles. Eur J Pharm Biopharmaceutics: Official J Arbeitsgemeinschaft fur Pharmazeutische Verfahrenstechnik eV. 2017;112:45–50. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

