Use of Genomics to Track Coronavirus Disease Outbreaks, New Zealand
- PMID: 33900175
- PMCID: PMC8084492
- DOI: 10.3201/eid2705.204579
Use of Genomics to Track Coronavirus Disease Outbreaks, New Zealand
Abstract
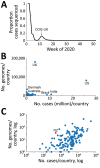
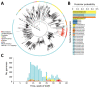
Real-time genomic sequencing has played a major role in tracking the global spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), contributing greatly to disease mitigation strategies. In August 2020, after having eliminated the virus, New Zealand experienced a second outbreak. During that outbreak, New Zealand used genomic sequencing in a primary role, leading to a second elimination of the virus. We generated genomes from 78% of the laboratory-confirmed samples of SARS-CoV-2 from the second outbreak and compared them with the available global genomic data. Genomic sequencing rapidly identified that virus causing the second outbreak in New Zealand belonged to a single cluster, thus resulting from a single introduction. However, successful identification of the origin of this outbreak was impeded by substantial biases and gaps in global sequencing data. Access to a broader and more heterogenous sample of global genomic data would strengthen efforts to locate the source of any new outbreaks.
Keywords: 2019 novel coronavirus disease; COVID-19; New Zealand; SARS-CoV-2; coronavirus disease; genomics; infectious disease; respiratory infections; severe acute respiratory syndrome coronavirus 2; viruses; zoonoses.
Figures
Similar articles
-
Real-Time Genomics for Tracking Severe Acute Respiratory Syndrome Coronavirus 2 Border Incursions after Virus Elimination, New Zealand.Emerg Infect Dis. 2021 Sep;27(9):2361-2368. doi: 10.3201/eid2709.211097. Emerg Infect Dis. 2021. PMID: 34424164 Free PMC article.
-
Whole-Genome Sequencing of SARS-CoV-2 from Quarantine Hotel Outbreak.Emerg Infect Dis. 2021 Aug;27(8):2219-2221. doi: 10.3201/eid2708.204875. Emerg Infect Dis. 2021. PMID: 34287141 Free PMC article.
-
Transmission of Severe Acute Respiratory Syndrome Coronavirus 2 during Border Quarantine and Air Travel, New Zealand (Aotearoa).Emerg Infect Dis. 2021 May;27(5):1274-1278. doi: 10.3201/eid2705.210514. Epub 2021 Mar 18. Emerg Infect Dis. 2021. PMID: 33734063 Free PMC article. Review.
-
Genomic epidemiology of Delta SARS-CoV-2 during transition from elimination to suppression in Aotearoa New Zealand.Nat Commun. 2022 Jul 12;13(1):4035. doi: 10.1038/s41467-022-31784-5. Nat Commun. 2022. PMID: 35821124 Free PMC article.
-
The Coronavirus Pandemic: What Does the Evidence Show?J Nepal Health Res Counc. 2020 Apr 19;18(1):1-9. doi: 10.33314/jnhrc.v18i1.2596. J Nepal Health Res Counc. 2020. PMID: 32335585 Review.
Cited by
-
In silico analyses identify sequence contamination thresholds for Nanopore-generated SARS-CoV-2 sequences.PLoS Comput Biol. 2024 Aug 19;20(8):e1011539. doi: 10.1371/journal.pcbi.1011539. eCollection 2024 Aug. PLoS Comput Biol. 2024. PMID: 39159257 Free PMC article.
-
Investigating the rise of Omicron variant through genomic surveillance of SARS-CoV-2 infections in a highly vaccinated university population.Microb Genom. 2024 Feb;10(2):001194. doi: 10.1099/mgen.0.001194. Microb Genom. 2024. PMID: 38334271 Free PMC article.
-
Phylodynamics reveals the role of human travel and contact tracing in controlling the first wave of COVID-19 in four island nations.Virus Evol. 2021 Jun 8;7(2):veab052. doi: 10.1093/ve/veab052. eCollection 2021. Virus Evol. 2021. PMID: 34527282 Free PMC article.
-
We should not dismiss the possibility of eradicating COVID-19: comparisons with smallpox and polio.BMJ Glob Health. 2021 Aug;6(8):e006810. doi: 10.1136/bmjgh-2021-006810. BMJ Glob Health. 2021. PMID: 34373261 Free PMC article. No abstract available.
-
Rapid Response to SARS-CoV-2 in Aotearoa New Zealand: Implementation of a Diagnostic Test and Characterization of the First COVID-19 Cases in the South Island.Viruses. 2021 Nov 4;13(11):2222. doi: 10.3390/v13112222. Viruses. 2021. PMID: 34835031 Free PMC article.
References
-
- Holmes EC. Novel 2019. coronavirus genome [cited 2021 Jan 20]. https://virological.org/t/novel-2019-coronavirus-genome/319
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

