Functional evaluation of conserved basic residues in human phosphomevalonate kinase
- PMID: 17902708
- PMCID: PMC2530820
- DOI: 10.1021/bi701408t
Functional evaluation of conserved basic residues in human phosphomevalonate kinase
Abstract
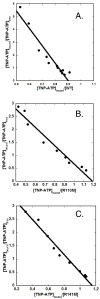
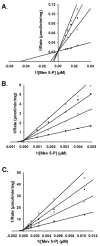
Phosphomevalonate kinase (PMK) catalyzes the cation-dependent reaction of mevalonate 5-phosphate with ATP to form mevalonate 5-diphosphate and ADP, a key step in the mevalonate pathway for isoprenoid/sterol biosynthesis. Animal PMK proteins belong to the nucleoside monophosphate (NMP) kinase family. For many NMP kinases, multiple basic residues contribute to the neutralization of the negatively charged pentacoordinate phosphate reaction intermediate. Loss of basicity can result in catalytically impaired enzymes. On the basis of this precedent, conserved basic residues of human PMK have been mutated, and purified forms of the mutated proteins have been kinetically and biophysically characterized. K48M and R73M mutants exhibit diminished Vmax values in both reaction directions (>1000-fold) with only slight Km perturbations (<10-fold). In both forward and reverse reactions, R110M exhibits a large (>10,000-fold) specific activity diminution. R111M exhibits substantially inflated Km values for mevalonate 5-phosphate and mevalonate 5-diphosphate (60- and 30-fold, respectively) as well as decreases [50-fold (forward) and 85-fold (reverse)] in Vmax. R84M also exhibits inflated Km values (50- and 33-fold for mevalonate 5-phosphate and mevalonate 5-diphosphate, respectively). The Ki values for R111M and R84M product inhibition by mevalonate 5-diphosphate are inflated by 45- and 63-fold; effects are comparable to the 30- and 38-fold inflations in Km for mevalonate 5-diphosphate. R141M exhibits little perturbation in Vmax [14-fold (forward) and 10-fold (reverse)] but has inflated Km values for ATP and ADP (48- and 136-fold, respectively). The Kd of ATP for R141M, determined by changes in tryptophan fluorescence, is inflated 27-fold compared to wt PMK. These data suggest that R110 is important to PMK catalysis, which is also influenced by K48 and R73. R111 and R84 contribute to binding of mevalonate 5-phosphate and R141 to binding of ATP.
Figures

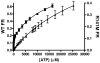
Similar articles
-
Phosphomevalonate kinase: functional investigation of the recombinant human enzyme.Biochemistry. 2006 Mar 14;45(10):3235-42. doi: 10.1021/bi052231u. Biochemistry. 2006. PMID: 16519518
-
Substrate induced structural and dynamics changes in human phosphomevalonate kinase and implications for mechanism.Proteins. 2009 Apr;75(1):127-38. doi: 10.1002/prot.22228. Proteins. 2009. PMID: 18798562 Free PMC article.
-
Investigation of the functional contributions of invariant serine residues in yeast mevalonate diphosphate decarboxylase.Biochemistry. 2005 Feb 22;44(7):2671-7. doi: 10.1021/bi0484217. Biochemistry. 2005. PMID: 15709780
-
Investigation of invariant serine/threonine residues in mevalonate kinase. Tests of the functional significance of a proposed substrate binding motif and a site implicated in human inherited disease.J Biol Chem. 2001 Apr 20;276(16):12573-8. doi: 10.1074/jbc.M011478200. Epub 2001 Jan 17. J Biol Chem. 2001. PMID: 11278915
-
Human mevalonate diphosphate decarboxylase: characterization, investigation of the mevalonate diphosphate binding site, and crystal structure.Arch Biochem Biophys. 2008 Dec 1;480(1):58-67. doi: 10.1016/j.abb.2008.08.024. Epub 2008 Sep 18. Arch Biochem Biophys. 2008. PMID: 18823933 Free PMC article.
Cited by
-
NMR dynamics investigation of ligand-induced changes of main and side-chain arginine N-H's in human phosphomevalonate kinase.J Am Chem Soc. 2010 Feb 24;132(7):2102-3. doi: 10.1021/ja906244j. J Am Chem Soc. 2010. PMID: 20112895 Free PMC article.
-
Second-Hit, Postzygotic PMVK and MVD Mutations in Linear Porokeratosis.JAMA Dermatol. 2019 May 1;155(5):548-555. doi: 10.1001/jamadermatol.2019.0016. JAMA Dermatol. 2019. PMID: 30942823 Free PMC article.
-
Molecular docking and NMR binding studies to identify novel inhibitors of human phosphomevalonate kinase.Biochem Biophys Res Commun. 2013 Jan 4;430(1):313-9. doi: 10.1016/j.bbrc.2012.10.130. Epub 2012 Nov 10. Biochem Biophys Res Commun. 2013. PMID: 23146631 Free PMC article.
-
Genome-wide association study for backfat thickness in Canchim beef cattle using Random Forest approach.BMC Genet. 2013 Jun 5;14:47. doi: 10.1186/1471-2156-14-47. BMC Genet. 2013. PMID: 23738659 Free PMC article.
-
A proteome-wide atlas of lysine-reactive chemistry.Nat Chem. 2021 Nov;13(11):1081-1092. doi: 10.1038/s41557-021-00765-4. Epub 2021 Sep 9. Nat Chem. 2021. PMID: 34504315 Free PMC article.
References
-
- Hellig H, Popjak G. Studies on the biosynthesis of cholesterol: XIII. phosphomevalonic kinase from liver. J Lipid Res. 1961;2:235–243.
-
- Bazaes S, Beytia E, Jabalquinto AM, Solis de Ovando F, Gomez I, Eyzaguirre J. Pig liver phosphomevalone kinase. 1. Purification and properties. Biochemistry. 1980;19:2300–2304. - PubMed
-
- Lee CS, O’Sullivan WJ. Improved procedures for the synthesis of phosphomevalonate and for the assay and purification of pig liver phosphomevalonate kinase. Biochim Biophys Acta. 1985;839:83–89. - PubMed
-
- Chambliss KL, Slaughter CA, Schreiner R, Hoffmann GF, Gibson KM. Molecular cloning of human phosphomevalonate kinase and identification of a consensus peroxisomal targeting sequence. J Biol Chem. 1996;271:17330–17334. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

