Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain
- PMID: 15774886
- PMCID: PMC1065214
- DOI: 10.1128/JB.187.7.2426-2438.2005
Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain
Abstract
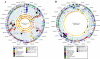
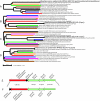
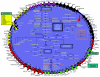
Staphylococcus aureus is an opportunistic pathogen and the major causative agent of numerous hospital- and community-acquired infections. Staphylococcus epidermidis has emerged as a causative agent of infections often associated with implanted medical devices. We have sequenced the approximately 2.8-Mb genome of S. aureus COL, an early methicillin-resistant isolate, and the approximately 2.6-Mb genome of S. epidermidis RP62a, a methicillin-resistant biofilm isolate. Comparative analysis of these and other staphylococcal genomes was used to explore the evolution of virulence and resistance between these two species. The S. aureus and S. epidermidis genomes are syntenic throughout their lengths and share a core set of 1,681 open reading frames. Genome islands in nonsyntenic regions are the primary source of variations in pathogenicity and resistance. Gene transfer between staphylococci and low-GC-content gram-positive bacteria appears to have shaped their virulence and resistance profiles. Integrated plasmids in S. epidermidis carry genes encoding resistance to cadmium and species-specific LPXTG surface proteins. A novel genome island encodes multiple phenol-soluble modulins, a potential S. epidermidis virulence factor. S. epidermidis contains the cap operon, encoding the polyglutamate capsule, a major virulence factor in Bacillus anthracis. Additional phenotypic differences are likely the result of single nucleotide polymorphisms, which are most numerous in cell envelope proteins. Overall differences in pathogenicity can be attributed to genome islands in S. aureus which encode enterotoxins, exotoxins, leukocidins, and leukotoxins not found in S. epidermidis.
Figures

Similar articles
-
Methicillin resistance transfer from Staphylocccus epidermidis to methicillin-susceptible Staphylococcus aureus in a patient during antibiotic therapy.PLoS One. 2010 Jul 29;5(7):e11841. doi: 10.1371/journal.pone.0011841. PLoS One. 2010. PMID: 20686601 Free PMC article.
-
Staphylococcus epidermidis MSCRAMM SesJ Is Encoded in Composite Islands.mBio. 2020 Feb 18;11(1):e02911-19. doi: 10.1128/mBio.02911-19. mBio. 2020. PMID: 32071265 Free PMC article.
-
Global acquisition of genetic material from different bacteria into the staphylococcal cassette chromosome elements of a Staphylococcus epidermidis isolate.Int J Antimicrob Agents. 2017 Oct;50(4):581-587. doi: 10.1016/j.ijantimicag.2017.06.015. Epub 2017 Jul 10. Int J Antimicrob Agents. 2017. PMID: 28705673
-
Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus.Lancet. 2006 Mar 4;367(9512):731-9. doi: 10.1016/S0140-6736(06)68231-7. Lancet. 2006. PMID: 16517273 Review.
-
Genomic variation and evolution of Staphylococcus aureus.Int J Med Microbiol. 2010 Feb;300(2-3):98-103. doi: 10.1016/j.ijmm.2009.08.013. Epub 2009 Oct 7. Int J Med Microbiol. 2010. PMID: 19811948 Review.
Cited by
-
Distribution of Staphylococcal Cassette Chromosome (SCC) mec Element Types in Fusidic Acid-Resistant Staphylococcus epidermidis and Identification of a Novel SCC7684 Element.Antimicrob Agents Chemother. 2016 Jul 22;60(8):5006-9. doi: 10.1128/AAC.00231-16. Print 2016 Aug. Antimicrob Agents Chemother. 2016. PMID: 27161629 Free PMC article.
-
Sodium Dodecyl Sulfate (SDS)-Loaded Nanoporous Polymer as Anti-Biofilm Surface Coating Material.Int J Mol Sci. 2013 Feb 1;14(2):3050-64. doi: 10.3390/ijms14023050. Int J Mol Sci. 2013. PMID: 23377015 Free PMC article.
-
Whole-Genome Analysis of Staphylococcus aureus Isolates from Ready-to-Eat Food in Russia.Foods. 2022 Aug 25;11(17):2574. doi: 10.3390/foods11172574. Foods. 2022. PMID: 36076759 Free PMC article.
-
Staphylococcus epidermidis pan-genome sequence analysis reveals diversity of skin commensal and hospital infection-associated isolates.Genome Biol. 2012 Jul 25;13(7):R64. doi: 10.1186/gb-2012-13-7-r64. Genome Biol. 2012. PMID: 22830599 Free PMC article.
-
Association of tannase-producing Staphylococcus lugdunensis with colon cancer and characterization of a novel tannase gene.J Gastroenterol. 2007 May;42(5):346-51. doi: 10.1007/s00535-007-2012-5. Epub 2007 May 25. J Gastroenterol. 2007. PMID: 17530358
References
-
- Archer, G. L., and D. M. Niemeyer. 1994. Origin and evolution of DNA associated with resistance to methicillin in staphylococci. Trends Microbiol. 2:343-347. - PubMed
-
- Baba, T., F. Takeuchi, M. Kuroda, H. Yuzawa, K. Aoki, A. Oguchi, Y. Nagai, N. Iwama, K. Asano, T. Naimi, H. Kuroda, L. Cui, K. Yamamoto, and K. Hiramatsu. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819-1827. - PubMed
-
- Beres, S. B., G. L. Sylva, K. D. Barbian, B. Lei, J. S. Hoff, N. D. Mammarella, M. Y. Liu, J. C. Smoot, S. F. Porcella, L. D. Parkins, D. S. Campbell, T. M. Smith, J. K. McCormick, D. Y. Leung, P. M. Schlievert, and J. M. Musser. 2002. Genome sequence of a serotype M3 strain of group A Streptococcus: phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proc. Natl. Acad. Sci. USA 99:10078-10083. - PMC - PubMed
-
- Centers for Disease Control and Prevention. 1999. Four pediatric deaths from community-acquired methicillin-resistant Staphylococcus aureus—Minnesota and North Dakota, 1997-1999. Morb. Mortal. Wkly. Rep. 48:707-710. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

