Structural insights into the diversity and DNA cleavage mechanism of Fanzor
- PMID: 39208796
- PMCID: PMC11423790
- DOI: 10.1016/j.cell.2024.07.050
Structural insights into the diversity and DNA cleavage mechanism of Fanzor
Abstract
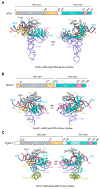
Fanzor (Fz) is an ωRNA-guided endonuclease extensively found throughout the eukaryotic domain with unique gene editing potential. Here, we describe the structures of Fzs from three different organisms. We find that Fzs share a common ωRNA interaction interface, regardless of the length of the ωRNA, which varies considerably across species. The analysis also reveals Fz's mode of DNA recognition and unwinding capabilities as well as the presence of a non-canonical catalytic site. The structures demonstrate how protein conformations of Fz shift to allow the binding of double-stranded DNA to the active site within the R-loop. Mechanistically, examination of structures in different states shows that the conformation of the lid loop on the RuvC domain is controlled by the formation of the guide/DNA heteroduplex, regulating the activation of nuclease and DNA double-stranded displacement at the single cleavage site. Our findings clarify the mechanism of Fz, establishing a foundation for engineering efforts.
Keywords: DNA cleavage mechanisms; Fanzor; R-loop structure; activation mechanisms; catalytic site; eukaryotic RNA-guided DNA endonuclease; gene editing; structural diversity.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests P.X., M.S., G.F., and F.Z. are coinventors on a patent application (PCT/US2022/081593) related to this work filed by the Broad Institute and MIT. F.Z. is a scientific advisor and cofounder of Editas Medicine, Beam Therapeutics, Pairwise Plants, Arbor Biotechnologies, Aera Therapeutics, and Moonwalk Biosciences. F.Z. is a scientific advisor for Octant.
Figures





Similar articles
-
Cas12a domain flexibility guides R-loop formation and forces RuvC resetting.Mol Cell. 2024 Jul 25;84(14):2717-2731.e6. doi: 10.1016/j.molcel.2024.06.007. Epub 2024 Jul 1. Mol Cell. 2024. PMID: 38955179
-
Structure of the mini-RNA-guided endonuclease CRISPR-Cas12j3.Nat Commun. 2021 Jul 22;12(1):4476. doi: 10.1038/s41467-021-24707-3. Nat Commun. 2021. PMID: 34294706 Free PMC article.
-
Mg2+-dependent conformational rearrangements of CRISPR-Cas12a R-loop complex are mandatory for complete double-stranded DNA cleavage.Proc Natl Acad Sci U S A. 2021 Dec 7;118(49):e2113747118. doi: 10.1073/pnas.2113747118. Proc Natl Acad Sci U S A. 2021. PMID: 34853172 Free PMC article.
-
CRISPR-Cas9 Structures and Mechanisms.Annu Rev Biophys. 2017 May 22;46:505-529. doi: 10.1146/annurev-biophys-062215-010822. Epub 2017 Mar 30. Annu Rev Biophys. 2017. PMID: 28375731 Review.
-
Editor's cut: DNA cleavage by CRISPR RNA-guided nucleases Cas9 and Cas12a.Biochem Soc Trans. 2020 Feb 28;48(1):207-219. doi: 10.1042/BST20190563. Biochem Soc Trans. 2020. PMID: 31872209 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

