Characterization of the dual regulation by a c-di-GMP riboswitch Bc1 with a long expression platform from Bacillus thuringiensis
- PMID: 38819160
- PMCID: PMC11218506
- DOI: 10.1128/spectrum.00450-24
Characterization of the dual regulation by a c-di-GMP riboswitch Bc1 with a long expression platform from Bacillus thuringiensis
Abstract
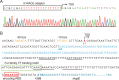
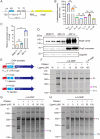
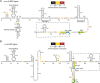
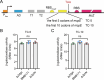
A riboswitch generally regulates the expression of its downstream genes through conformational change in its expression platform (EP) upon ligand binding. The cyclic diguanosine monophosphate (c-di-GMP) class I riboswitch Bc1 is widespread and conserved among Bacillus cereus group species. In this study, we revealed that Bc1 has a long EP with two typical ρ-independent terminator sequences 28 bp apart. The upstream terminator T1 is dominant in vitro, while downstream terminator T2 is more efficient in vivo. Through mutation analysis, we elucidated that Bc1 exerts a rare and incoherent "transcription-translation" dual regulation with T2 playing a crucial role. However, we found that Bc1 did not respond to c-di-GMP under in vitro transcription conditions, and the expressions of downstream genes did not change with fluctuation in intracellular c-di-GMP concentration. To explore this puzzle, we conducted SHAPE-MaP and confirmed the interaction of Bc1 with c-di-GMP. This shows that as c-di-GMP concentration increases, T1 unfolds but T2 remains almost intact and functional. The presence of T2 masks the effect of T1 unwinding, resulting in no response of Bc1 to c-di-GMP. The high Shannon entropy values of EP region imply the potential alternative structures of Bc1. We also found that zinc uptake regulator can specifically bind to the dual terminator coding sequence and slightly trigger the response of Bc1 to c-di-GMP. This work will shed light on the dual-regulation riboswitch and enrich our understanding of the RNA world.IMPORTANCEIn nature, riboswitches are involved in a variety of metabolic regulation, most of which preferentially regulate transcription termination or translation initiation of downstream genes in specific ways. Alternatively, the same or different riboswitches can exist in tandem to enhance regulatory effects or respond to multiple ligands. However, many putative conserved riboswitches have not yet been experimentally validated. Here, we found that the c-di-GMP riboswitch Bc1 with a long EP could form a dual terminator and exhibit non-canonical and incoherent "transcription-translation" dual regulation. Besides, zinc uptake regulator specifically bound to the coding sequence of the Bc1 EP and slightly mediated the action of Bc1. The application of SHAPE-MaP to the dual regulation mechanism of Bc1 may establish the foundation for future studies of such complex untranslated regions in other bacterial genomes.
Keywords: Bacillus cereus group; SHAPE-MaP; Zur (zinc uptake regulator); cyclic di-GMP riboswitch; dual regulation; dual terminator.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
The Vc2 Cyclic di-GMP-Dependent Riboswitch of Vibrio cholerae Regulates Expression of an Upstream Putative Small RNA by Controlling RNA Stability.J Bacteriol. 2019 Oct 4;201(21):e00293-19. doi: 10.1128/JB.00293-19. Print 2019 Nov 1. J Bacteriol. 2019. PMID: 31405916 Free PMC article.
-
Cyclic di-GMP riboswitch-regulated type IV pili contribute to aggregation of Clostridium difficile.J Bacteriol. 2015 Mar;197(5):819-32. doi: 10.1128/JB.02340-14. Epub 2014 Dec 15. J Bacteriol. 2015. PMID: 25512308 Free PMC article.
-
[Structure and function of c-di-GMP riboswitches].Sheng Wu Gong Cheng Xue Bao. 2017 Sep 25;33(9):1357-1368. doi: 10.13345/j.cjb.170085. Sheng Wu Gong Cheng Xue Bao. 2017. PMID: 28956387 Review. Chinese.
-
Characterization of a natural triple-tandem c-di-GMP riboswitch and application of the riboswitch-based dual-fluorescence reporter.Sci Rep. 2016 Feb 19;6:20871. doi: 10.1038/srep20871. Sci Rep. 2016. PMID: 26892868 Free PMC article.
-
You've come a long way: c-di-GMP signaling.Curr Opin Microbiol. 2012 Apr;15(2):140-6. doi: 10.1016/j.mib.2011.12.008. Epub 2012 Jan 5. Curr Opin Microbiol. 2012. PMID: 22226607 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

