Bioinformatic and functional characterization of cyclic-di-GMP metabolic proteins in Vibrio alginolyticus unveils key diguanylate cyclases controlling multiple biofilm-associated phenotypes
- PMID: 37808288
- PMCID: PMC10552763
- DOI: 10.3389/fmicb.2023.1258415
Bioinformatic and functional characterization of cyclic-di-GMP metabolic proteins in Vibrio alginolyticus unveils key diguanylate cyclases controlling multiple biofilm-associated phenotypes
Abstract
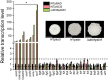
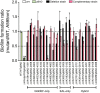
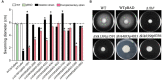
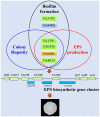
The biofilm lifestyle is critical for bacterial survival and proliferation in the fluctuating marine environment. Cyclic diguanylate (c-di-GMP) is a key second messenger during bacterial adaptation to various environmental signals, which has been identified as a master regulator of biofilm formation. However, little is known about whether and how c-di-GMP signaling regulates biofilm formation in Vibrio alginolyticus, a globally dominant marine pathogen. Here, a large set of 63 proteins were predicted to participate in c-di-GMP metabolism (biosynthesis or degradation) in a pathogenic V. alginolyticus strain HN08155. Guided by protein homology, conserved domains and gene context information, a representative subset of 22 c-di-GMP metabolic proteins were selected to determine which ones affect biofilm-associated phenotypes. By comparing phenotypic differences between the wild-type and mutants or overexpression strains, we found that 22 c-di-GMP metabolic proteins can separately regulate different phenotypic outputs in V. alginolyticus. The results indicated that overexpression of four c-di-GMP metabolic proteins, including VA0356, VA1591 (CdgM), VA4033 (DgcB) and VA0088, strongly enhanced rugose colony morphotypes and strengthened Congo Red (CR) binding capacity, both of which are indicators of biofilm matrix overproduction. Furthermore, rugose enhanced colonies were accompanied by increased transcript levels of extracellular polysaccharide (EPS) biosynthesis genes and decreased expression of flagellar synthesis genes compared to smooth colonies (WTpBAD control), as demonstrated by overexpression strains WTp4033 and ∆VA4033p4033. Overall, the high abundance of c-di-GMP metabolic proteins in V. alginolyticus suggests that c-di-GMP signaling and regulatory system could play a key role in its response and adaptation to the ever-changing marine environment. This work provides a robust foundation for the study of the molecular mechanisms of c-di-GMP in the biofilm formation of V. alginolyticus.
Keywords: Vibrio alginolyticus; biofilm; c-di-GMP; extracellular polysaccharide; rugose.
Copyright © 2023 Gong, Zeng, Chen, Zhang, Han, Long and Xie.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Environmental Calcium Initiates a Feed-Forward Signaling Circuit That Regulates Biofilm Formation and Rugosity in Vibrio vulnificus.mBio. 2018 Aug 28;9(4):e01377-18. doi: 10.1128/mBio.01377-18. mBio. 2018. PMID: 30154262 Free PMC article.
-
Para-Aminobenzoic Acid, Calcium, and c-di-GMP Induce Formation of Cohesive, Syp-Polysaccharide-Dependent Biofilms in Vibrio fischeri.mBio. 2021 Oct 26;12(5):e0203421. doi: 10.1128/mBio.02034-21. Epub 2021 Oct 5. mBio. 2021. PMID: 34607467 Free PMC article.
-
Functional Specialization in Vibrio cholerae Diguanylate Cyclases: Distinct Modes of Motility Suppression and c-di-GMP Production.mBio. 2019 Apr 23;10(2):e00670-19. doi: 10.1128/mBio.00670-19. mBio. 2019. PMID: 31015332 Free PMC article.
-
Diguanylate Cyclases in Vibrio cholerae: Essential Regulators of Lifestyle Switching.Front Cell Infect Microbiol. 2020 Oct 22;10:582947. doi: 10.3389/fcimb.2020.582947. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 33194821 Free PMC article. Review.
-
Bacterial diguanylate cyclases: structure, function and mechanism in exopolysaccharide biofilm development.Biotechnol Adv. 2015 Jan-Feb;33(1):124-141. doi: 10.1016/j.biotechadv.2014.11.010. Epub 2014 Dec 10. Biotechnol Adv. 2015. PMID: 25499693 Review.
References
LinkOut - more resources
Full Text Sources

