Bioinformatic analysis of the LCN2-SLC22A17-MMP9 network in cancer: The role of DNA methylation in the modulation of tumor microenvironment
- PMID: 36211450
- PMCID: PMC9532607
- DOI: 10.3389/fcell.2022.945586
Bioinformatic analysis of the LCN2-SLC22A17-MMP9 network in cancer: The role of DNA methylation in the modulation of tumor microenvironment
Abstract
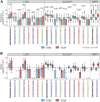
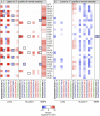
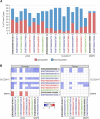
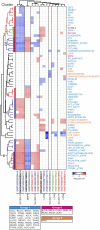
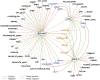
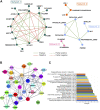
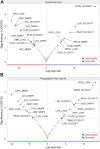
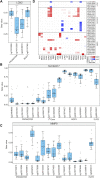
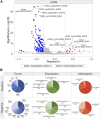
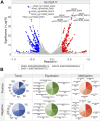
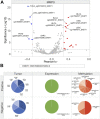
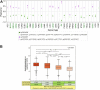
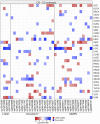
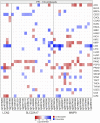
Several features of cancer cells such as proliferation, invasion, metastatic spreading, and drug resistance are affected by their interaction with several tumor microenvironment (TME) components, including neutrophil gelatinase-associated lipocalin (NGAL), solute carrier family 22 member 17 (SLC22A17), and matrix metallopeptidase 9 (MMP9). These molecules play a key role in tumor growth, invasion, and iron-dependent metabolism of cancer cells. However, the precise epigenetic mechanisms underlying the gene regulation of Lipocalin 2 (LCN2), SLC22A17, and MMP9 in cancer still remain unclear. To this purpose, computational analysis was performed on TCGA and GTEx datasets to evaluate the expression and DNA methylation status of LCN2, SLC22A17, and MMP9 genes in different tumor types. Correlation analysis between gene/isoforms expression and DNA methylation levels of LCN2, SLC22A17, and MMP9 was performed to investigate the role of DNA methylation in the modulation of these genes. Protein network analysis was carried out using reverse phase protein arrays (RPPA) data to identify protein-protein interactions of the LCN2-SLC22A17-MMP9 network. Furthermore, survival analysis was performed according to gene expression and DNA methylation levels. Our results demonstrated that LCN2 and MMP9 were mainly upregulated in most tumor types, whereas SLC22A17 was largely downregulated, representing a specific hallmark signature for all gastrointestinal tumors. Notably, the expression of LCN2, SLC22A17, and MMP9 genes was negatively affected by promoter methylation. Conversely, intragenic hypermethylation was associated with the overexpression of SLC22A17 and MMP9 genes. Protein network analysis highlighted the role of the LCN2-SLC22A17-MMP9 network in TME by the interaction with fibronectin 1 and claudin 7, especially in rectal tumors. Moreover, the impact of expression and methylation status of LCN2, SLC22A17, and MMP9 on overall survival and progression free interval was tumor type-dependent. Overall, our analyses provide a detailed overview of the expression and methylation status of LCN2, SLC22A17, and MMP9 in all TCGA tumors, indicating that the LCN2-SLC22A17-MMP9 network was strictly regulated by DNA methylation within TME. Our findings pave the way for the identification of novel DNA methylation hotspots with diagnostic and prognostic values and suitable for epi-drug targeting.
Keywords: DNA methylation; LCN2; MMP9; SLC22A17; bioinformatic; cancer; ferroptosis; gene expression.
Copyright © 2022 Candido, Tomasello, Lavoro, Falzone, Gattuso, Russo, Paratore, McCubrey and Libra.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Identification of SLC22A17 DNA methylation hotspot as a potential biomarker in cutaneous melanoma.J Transl Med. 2024 Oct 2;22(1):887. doi: 10.1186/s12967-024-05622-9. J Transl Med. 2024. PMID: 39358721 Free PMC article.
-
Immunohistochemical detection of a specific receptor for lipocalin2 (solute carrier family 22 member 17, SLC22A17) and its prognostic significance in endometrial carcinoma.Exp Mol Pathol. 2011 Oct;91(2):563-8. doi: 10.1016/j.yexmp.2011.06.002. Epub 2011 Jul 8. Exp Mol Pathol. 2011. PMID: 21763306
-
Inverse Regulation of Lipocalin-2/24p3 Receptor/SLC22A17 and Lipocalin-2 Expression by Tonicity, NFAT5/TonEBP and Arginine Vasopressin in Mouse Cortical Collecting Duct Cells mCCD(cl.1): Implications for Osmotolerance.Int J Mol Sci. 2019 Oct 30;20(21):5398. doi: 10.3390/ijms20215398. Int J Mol Sci. 2019. PMID: 31671521 Free PMC article.
-
Role of the SLC22A17/lipocalin-2 receptor in renal endocytosis of proteins/metalloproteins: a focus on iron- and cadmium-binding proteins.Am J Physiol Renal Physiol. 2023 Nov 1;325(5):F564-F577. doi: 10.1152/ajprenal.00020.2023. Epub 2023 Aug 17. Am J Physiol Renal Physiol. 2023. PMID: 37589051 Review.
-
Implication and role of neutrophil gelatinase-associated lipocalin in cancer: lipocalin-2 as a potential novel emerging comprehensive therapeutic target for a variety of cancer types.Mol Biol Rep. 2020 Mar;47(3):2327-2346. doi: 10.1007/s11033-020-05261-5. Epub 2020 Jan 22. Mol Biol Rep. 2020. PMID: 31970626 Review.
Cited by
-
Natural Compounds and Glutathione: Beyond Mere Antioxidants.Antioxidants (Basel). 2023 Jul 18;12(7):1445. doi: 10.3390/antiox12071445. Antioxidants (Basel). 2023. PMID: 37507985 Free PMC article. Review.
-
Identification of miRNAs in extracellular vesicles as potential diagnostic markers for pediatric epilepsy and drug-resistant epilepsy via bioinformatics analysis.Front Pediatr. 2023 Jul 3;11:1199780. doi: 10.3389/fped.2023.1199780. eCollection 2023. Front Pediatr. 2023. PMID: 37469680 Free PMC article.
-
Identification of SLC22A17 DNA methylation hotspot as a potential biomarker in cutaneous melanoma.J Transl Med. 2024 Oct 2;22(1):887. doi: 10.1186/s12967-024-05622-9. J Transl Med. 2024. PMID: 39358721 Free PMC article.
-
Lipocalin-2: A Nurturer of Tumor Progression and a Novel Candidate for Targeted Cancer Therapy.Cancers (Basel). 2023 Oct 26;15(21):5159. doi: 10.3390/cancers15215159. Cancers (Basel). 2023. PMID: 37958332 Free PMC article. Review.
-
Methylation‑sensitive restriction enzyme‑droplet digital PCR assay for the one‑step highly sensitive analysis of DNA methylation hotspots.Int J Mol Med. 2024 May;53(5):42. doi: 10.3892/ijmm.2024.5366. Epub 2024 Mar 15. Int J Mol Med. 2024. PMID: 38488030 Free PMC article.
References
-
- Abdelsameea E., Nada A., Omar N., Saleh S. M., Naguib M., El-Ezawy H. E. M., et al. (2020). Urine neutrophil gelatinase-associated lipocalin a possible diagnostic marker for Egyptian hepatocellular carcinoma patients. Asian pac. J. Cancer Prev. 21, 2259–2264. 10.31557/APJCP.2020.21.8.2259 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

