Dichloromethane Degradation Pathway from Unsequenced Hyphomicrobium sp. MC8b Rapidly Explored by Pan-Proteomics
- PMID: 33260855
- PMCID: PMC7760279
- DOI: 10.3390/microorganisms8121876
Dichloromethane Degradation Pathway from Unsequenced Hyphomicrobium sp. MC8b Rapidly Explored by Pan-Proteomics
Abstract
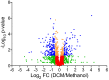
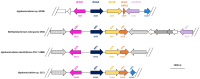
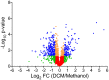
Several bacteria are able to degrade the major industrial solvent dichloromethane (DCM) by using the conserved dehalogenase DcmA, the only system for DCM degradation characterised at the sequence level so far. Using differential proteomics, we rapidly identified key determinants of DCM degradation for Hyphomicrobium sp. MC8b, an unsequenced facultative methylotrophic DCM-degrading strain. For this, we designed a pan-proteomics database comprising the annotated genome sequences of 13 distinct Hyphomicrobium strains. Compared to growth with methanol, growth with DCM induces drastic changes in the proteome of strain MC8b. Dichloromethane dehalogenase DcmA was detected by differential pan-proteomics, but only with poor sequence coverage, suggesting atypical characteristics of the DCM dehalogenation system in this strain. More peptides were assigned to DcmA by error-tolerant search, warranting subsequent sequencing of the genome of strain MC8b, which revealed a highly divergent set of dcm genes in this strain. This suggests that the dcm enzymatic system is less strongly conserved than previously believed, and that substantial molecular evolution of dcm genes has occurred beyond their horizontal transfer in the bacterial domain. Our study showed the power of pan-proteomics for quick characterization of new strains belonging to branches of the Tree of Life that are densely genome-sequenced.
Keywords: Hyphomicrobium; Nanopore; dcm genes; dehalogenation; dichloromethane; differential proteomics; genome sequencing; pan-proteomics.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures


Similar articles
-
Molecular characterization of dichloromethane-degrading Hyphomicrobium strains using 16S rDNA and DCM dehalogenase gene sequences.Syst Appl Microbiol. 2005 Sep;28(7):582-7. doi: 10.1016/j.syapm.2005.03.011. Syst Appl Microbiol. 2005. PMID: 16156115
-
Sequence variation in dichloromethane dehalogenases/glutathione S-transferases.Microbiology (Reading). 2001 Mar;147(Pt 3):611-619. doi: 10.1099/00221287-147-3-611. Microbiology (Reading). 2001. PMID: 11238968
-
Functional genomics of dichloromethane utilization in Methylobacterium extorquens DM4.Environ Microbiol. 2011 Sep;13(9):2518-35. doi: 10.1111/j.1462-2920.2011.02524.x. Epub 2011 Aug 19. Environ Microbiol. 2011. PMID: 21854516
-
Microbes, enzymes and genes involved in dichloromethane utilization.Biodegradation. 1994 Dec;5(3-4):237-48. doi: 10.1007/BF00696462. Biodegradation. 1994. PMID: 7765835 Review.
-
Bacterial growth with chlorinated methanes.Environ Health Perspect. 1995 Jun;103 Suppl 5(Suppl 5):33-6. doi: 10.1289/ehp.95103s433. Environ Health Perspect. 1995. PMID: 8565906 Free PMC article. Review.
Cited by
-
Characterizing the Microbial Consortium L1 Capable of Efficiently Degrading Chlorimuron-Ethyl via Metagenome Combining 16S rDNA Sequencing.Front Microbiol. 2022 Jun 23;13:912312. doi: 10.3389/fmicb.2022.912312. eCollection 2022. Front Microbiol. 2022. PMID: 35814706 Free PMC article.
-
Investigating the degradation potential of microbial consortia for perfluorooctane sulfonate through a functional "top-down" screening approach.PLoS One. 2024 May 17;19(5):e0303904. doi: 10.1371/journal.pone.0303904. eCollection 2024. PLoS One. 2024. PMID: 38758752 Free PMC article.
-
MecE, MecB, and MecC proteins orchestrate methyl group transfer during dichloromethane fermentation.Appl Environ Microbiol. 2024 Oct 23;90(10):e0097824. doi: 10.1128/aem.00978-24. Epub 2024 Sep 25. Appl Environ Microbiol. 2024. PMID: 39320083
-
Methylotrophic bacteria from rice paddy soils: mineral-nitrogen-utilizing isolates richness in bulk soil and rhizosphere.World J Microbiol Biotechnol. 2024 May 4;40(6):188. doi: 10.1007/s11274-024-04000-3. World J Microbiol Biotechnol. 2024. PMID: 38702590
References
-
- Bull A.T. Microbial Diversity and Bioprospecting. American Society of Microbiology; Washington, DC, USA: 2004. - DOI
-
- Silva W.M., Sousa C.S., Oliveira L.C., Soares S.C., Souza G.F.M.H., Tavares G.C., Resende C.P., Folador E.L., Pereira F.L., Figueiredo H., et al. Comparative proteomic analysis of four biotechnological strains Lactococcus lactis through label-free quantitative proteomics. Microb. Biotechnol. 2019;12:265–274. doi: 10.1111/1751-7915.13305. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

