Ecological drivers of bacterial community assembly in synthetic phycospheres
- PMID: 32015111
- PMCID: PMC7035482
- DOI: 10.1073/pnas.1917265117
Ecological drivers of bacterial community assembly in synthetic phycospheres
Abstract
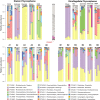
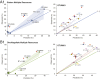
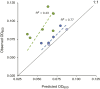
In the nutrient-rich region surrounding marine phytoplankton cells, heterotrophic bacterioplankton transform a major fraction of recently fixed carbon through the uptake and catabolism of phytoplankton metabolites. We sought to understand the rules by which marine bacterial communities assemble in these nutrient-enhanced phycospheres, specifically addressing the role of host resources in driving community coalescence. Synthetic systems with varying combinations of known exometabolites of marine phytoplankton were inoculated with seawater bacterial assemblages, and communities were transferred daily to mimic the average duration of natural phycospheres. We found that bacterial community assembly was predictable from linear combinations of the taxa maintained on each individual metabolite in the mixture, weighted for the growth each supported. Deviations from this simple additive resource model were observed but also attributed to resource-based factors via enhanced bacterial growth when host metabolites were available concurrently. The ability of photosynthetic hosts to shape bacterial associates through excreted metabolites represents a mechanism by which microbiomes with beneficial effects on host growth could be recruited. In the surface ocean, resource-based assembly of host-associated communities may underpin the evolution and maintenance of microbial interactions and determine the fate of a substantial portion of Earth's primary production.
Keywords: community assembly; phycospheres; phytoplankton–bacteria interactions.
Copyright © 2020 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures

Similar articles
-
Sulfonate-based networks between eukaryotic phytoplankton and heterotrophic bacteria in the surface ocean.Nat Microbiol. 2019 Oct;4(10):1706-1715. doi: 10.1038/s41564-019-0507-5. Epub 2019 Jul 22. Nat Microbiol. 2019. PMID: 31332382
-
Bacterial alkylquinolone signaling contributes to structuring microbial communities in the ocean.Microbiome. 2019 Jun 17;7(1):93. doi: 10.1186/s40168-019-0711-9. Microbiome. 2019. PMID: 31208456 Free PMC article.
-
Metagenomic and Metaproteomic Insights into Photoautotrophic and Heterotrophic Interactions in a Synechococcus Culture.mBio. 2020 Feb 18;11(1):e03261-19. doi: 10.1128/mBio.03261-19. mBio. 2020. PMID: 32071270 Free PMC article.
-
A multiomics approach to study the microbiome response to phytoplankton blooms.Appl Microbiol Biotechnol. 2017 Jun;101(12):4863-4870. doi: 10.1007/s00253-017-8330-5. Epub 2017 May 19. Appl Microbiol Biotechnol. 2017. PMID: 28526980 Review.
-
Space, the final frontier: The spatial component of phytoplankton-bacterial interactions.Mol Microbiol. 2024 Sep;122(3):331-346. doi: 10.1111/mmi.15293. Epub 2024 Jul 6. Mol Microbiol. 2024. PMID: 38970428 Review.
Cited by
-
Illuminating the dark metabolome of Pseudo-nitzschia-microbiome associations.Environ Microbiol. 2022 Nov;24(11):5408-5424. doi: 10.1111/1462-2920.16242. Environ Microbiol. 2022. PMID: 36222155 Free PMC article.
-
Changes in Cyanobacterial Phytoplankton Communities in Lake-Water Mesocosms Treated with Either Glucose or Hydrogen Peroxide.Microorganisms. 2024 Sep 22;12(9):1925. doi: 10.3390/microorganisms12091925. Microorganisms. 2024. PMID: 39338598 Free PMC article.
-
Experimental and computational approaches to unravel microbial community assembly.Comput Struct Biotechnol J. 2020 Dec 3;18:4071-4081. doi: 10.1016/j.csbj.2020.11.031. eCollection 2020. Comput Struct Biotechnol J. 2020. PMID: 33363703 Free PMC article. Review.
-
A Model Roseobacter, Ruegeria pomeroyi DSS-3, Employs a Diffusible Killing Mechanism To Eliminate Competitors.mSystems. 2020 Aug 11;5(4):e00443-20. doi: 10.1128/mSystems.00443-20. mSystems. 2020. PMID: 32788406 Free PMC article.
-
Host genotype structures the microbiome of a globally dispersed marine phytoplankton.Proc Natl Acad Sci U S A. 2021 Nov 30;118(48):e2105207118. doi: 10.1073/pnas.2105207118. Proc Natl Acad Sci U S A. 2021. PMID: 34810258 Free PMC article.
References
-
- Azam F., et al. , The ecological role of water-column microbes. Mar. Ecol. Prog. Ser. 10, 257–263 (1983).
-
- Cole J. J., Findlay S., Pace M. L., Bacterial production in fresh and saltwater ecosystems: A cross-system overview. Mar. Ecol. Prog. Ser. 43, 1–10 (1988).
-
- Thornton D. C., Dissolved organic matter (DOM) release by phytoplankton in the contemporary and future ocean. Eur. J. Phycol. 49, 20–46 (2014).
-
- Seymour J. R., Amin S. A., Raina J.-B., Stocker R., Zooming in on the phycosphere: The ecological interface for phytoplankton-bacteria relationships. Nat. Microbiol. 2, 17065 (2017). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

