Rates, distribution and implications of postzygotic mosaic mutations in autism spectrum disorder
- PMID: 28714951
- PMCID: PMC5672813
- DOI: 10.1038/nn.4598
Rates, distribution and implications of postzygotic mosaic mutations in autism spectrum disorder
Erratum in
-
Publisher Correction: Rates, distribution and implications of postzygotic mosaic mutations in autism spectrum disorder.Nat Neurosci. 2020 Sep;23(9):1176. doi: 10.1038/s41593-020-0681-z. Nat Neurosci. 2020. PMID: 32665711
Abstract
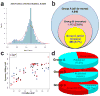
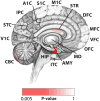
We systematically analyzed postzygotic mutations (PZMs) in whole-exome sequences from the largest collection of trios (5,947) with autism spectrum disorder (ASD) available, including 282 unpublished trios, and performed resequencing using multiple independent technologies. We identified 7.5% of de novo mutations as PZMs, 83.3% of which were not described in previous studies. Damaging, nonsynonymous PZMs within critical exons of prenatally expressed genes were more common in ASD probands than controls (P < 1 × 10-6), and genes carrying these PZMs were enriched for expression in the amygdala (P = 5.4 × 10-3). Two genes (KLF16 and MSANTD2) were significantly enriched for PZMs genome-wide, and other PZMs involved genes (SCN2A, HNRNPU and SMARCA4) whose mutation is known to cause ASD or other neurodevelopmental disorders. PZMs constitute a significant proportion of de novo mutations and contribute importantly to ASD risk.
Figures

Similar articles
-
Exonic Mosaic Mutations Contribute Risk for Autism Spectrum Disorder.Am J Hum Genet. 2017 Sep 7;101(3):369-390. doi: 10.1016/j.ajhg.2017.07.016. Epub 2017 Aug 31. Am J Hum Genet. 2017. PMID: 28867142 Free PMC article.
-
Exploring the biological role of postzygotic and germinal de novo mutations in ASD.Sci Rep. 2021 Jan 11;11(1):319. doi: 10.1038/s41598-020-79412-w. Sci Rep. 2021. PMID: 33431980 Free PMC article.
-
Postzygotic single-nucleotide mosaicisms contribute to the etiology of autism spectrum disorder and autistic traits and the origin of mutations.Hum Mutat. 2017 Aug;38(8):1002-1013. doi: 10.1002/humu.23255. Epub 2017 May 30. Hum Mutat. 2017. PMID: 28503910 Free PMC article.
-
Somatic Mosaicism and Autism Spectrum Disorder.Genes (Basel). 2021 Oct 26;12(11):1699. doi: 10.3390/genes12111699. Genes (Basel). 2021. PMID: 34828306 Free PMC article. Review.
-
Genetics of autism spectrum disorder.Handb Clin Neurol. 2018;147:321-329. doi: 10.1016/B978-0-444-63233-3.00021-X. Handb Clin Neurol. 2018. PMID: 29325621 Review.
Cited by
-
Tanc2-mediated mTOR inhibition balances mTORC1/2 signaling in the developing mouse brain and human neurons.Nat Commun. 2021 May 11;12(1):2695. doi: 10.1038/s41467-021-22908-4. Nat Commun. 2021. PMID: 33976205 Free PMC article.
-
De novo mutations, genetic mosaicism and human disease.Front Genet. 2022 Sep 26;13:983668. doi: 10.3389/fgene.2022.983668. eCollection 2022. Front Genet. 2022. PMID: 36226191 Free PMC article. Review.
-
Molecular Basis of the Schuurs-Hoeijmakers Syndrome: What We Know about the Gene and the PACS-1 Protein and Novel Therapeutic Approaches.Int J Mol Sci. 2022 Aug 25;23(17):9649. doi: 10.3390/ijms23179649. Int J Mol Sci. 2022. PMID: 36077045 Free PMC article. Review.
-
Next-Generation Sequencing in Autism Spectrum Disorder.Cold Spring Harb Perspect Med. 2019 Aug 1;9(8):a026872. doi: 10.1101/cshperspect.a026872. Cold Spring Harb Perspect Med. 2019. PMID: 30420340 Free PMC article. Review.
-
The state of research on the genetics of autism spectrum disorder: methodological, clinical and conceptual progress.Curr Opin Psychol. 2019 Jun;27:1-5. doi: 10.1016/j.copsyc.2018.07.004. Epub 2018 Jul 21. Curr Opin Psychol. 2019. PMID: 30059871 Free PMC article. Review.
References
MeSH terms
Grants and funding
- R01 MH097849/MH/NIMH NIH HHS/United States
- RC2 MH089952/MH/NIMH NIH HHS/United States
- S10 OD018522/OD/NIH HHS/United States
- RM1 HG008525/HG/NHGRI NIH HHS/United States
- RC2 HL102924/HL/NHLBI NIH HHS/United States
- U01 MH111662/MH/NIMH NIH HHS/United States
- U54 HD090255/HD/NICHD NIH HHS/United States
- RC2 HL103010/HL/NHLBI NIH HHS/United States
- R01 MH095797/MH/NIMH NIH HHS/United States
- S10 RR028832/RR/NCRR NIH HHS/United States
- R01 NS032457/NS/NINDS NIH HHS/United States
- R01 MH083565/MH/NIMH NIH HHS/United States
- RC2 HL102923/HL/NHLBI NIH HHS/United States
- UC2 HL102926/HL/NHLBI NIH HHS/United States
- R01 MH106910/MH/NIMH NIH HHS/United States
- UC2 HL103010/HL/NHLBI NIH HHS/United States
- U01 MH106883/MH/NIMH NIH HHS/United States
- T32 GM007226/GM/NIGMS NIH HHS/United States
- U01 MH100229/MH/NIMH NIH HHS/United States
- RC2 HL102926/HL/NHLBI NIH HHS/United States
- P50 HG005550/HG/NHGRI NIH HHS/United States
- U01 MH100209/MH/NIMH NIH HHS/United States
- U01 MH100239/MH/NIMH NIH HHS/United States
- T32 GM007753/GM/NIGMS NIH HHS/United States
- U01 MH111661/MH/NIMH NIH HHS/United States
- R21 MH106888/MH/NIMH NIH HHS/United States
- UC2 HL102923/HL/NHLBI NIH HHS/United States
- UC2 HL102924/HL/NHLBI NIH HHS/United States
- U01 MH100233/MH/NIMH NIH HHS/United States
- R01 MH113362/MH/NIMH NIH HHS/United States
- RC2 HL102925/HL/NHLBI NIH HHS/United States
- UC2 HL102925/HL/NHLBI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

