Mutations in mitochondrial DNA causing tubulointerstitial kidney disease
- PMID: 28267784
- PMCID: PMC5360345
- DOI: 10.1371/journal.pgen.1006620
Mutations in mitochondrial DNA causing tubulointerstitial kidney disease
Abstract
Tubulointerstitial kidney disease is an important cause of progressive renal failure whose aetiology is incompletely understood. We analysed a large pedigree with maternally inherited tubulointerstitial kidney disease and identified a homoplasmic substitution in the control region of the mitochondrial genome (m.547A>T). While mutations in mtDNA coding sequence are a well recognised cause of disease affecting multiple organs, mutations in the control region have never been shown to cause disease. Strikingly, our patients did not have classical features of mitochondrial disease. Patient fibroblasts showed reduced levels of mitochondrial tRNAPhe, tRNALeu1 and reduced mitochondrial protein translation and respiration. Mitochondrial transfer demonstrated mitochondrial transmission of the defect and in vitro assays showed reduced activity of the heavy strand promoter. We also identified further kindreds with the same phenotype carrying a homoplasmic mutation in mitochondrial tRNAPhe (m.616T>C). Thus mutations in mitochondrial DNA can cause maternally inherited renal disease, likely mediated through reduced function of mitochondrial tRNAPhe.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

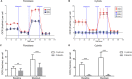
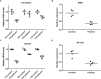
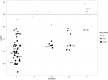


Similar articles
-
Homoplasmy of the Mitochondrial DNA Mutation m.616T>C Leads to Mitochondrial Tubulointerstitial Kidney Disease and Encephalopathia.Nephron. 2020;144(3):156-160. doi: 10.1159/000504412. Epub 2019 Nov 13. Nephron. 2020. PMID: 31722346
-
Gitelman-Like Syndrome Caused by Pathogenic Variants in mtDNA.J Am Soc Nephrol. 2022 Feb;33(2):305-325. doi: 10.1681/ASN.2021050596. Epub 2021 Oct 4. J Am Soc Nephrol. 2022. PMID: 34607911 Free PMC article.
-
Functional and morphological abnormalities of mitochondria harbouring the tRNA(Leu)(UUR) mutation in mitochondrial DNA derived from patients with maternally inherited diabetes and deafness (MIDD) and progressive kidney disease.Diabetologia. 1999 Apr;42(4):485-92. doi: 10.1007/s001250051183. Diabetologia. 1999. PMID: 10230654
-
Genetic Counselling for Maternally Inherited Mitochondrial Disorders.Mol Diagn Ther. 2017 Aug;21(4):419-429. doi: 10.1007/s40291-017-0279-7. Mol Diagn Ther. 2017. PMID: 28536827 Review.
-
Mitochondrial DNA mutations in human degenerative diseases and aging.Biochim Biophys Acta. 1995 May 24;1271(1):141-51. doi: 10.1016/0925-4439(95)00021-u. Biochim Biophys Acta. 1995. PMID: 7599200 Review.
Cited by
-
Mitophagy, Inflammasomes and Their Interaction in Kidney Diseases: A Comprehensive Review of Experimental Studies.J Inflamm Res. 2023 Apr 5;16:1457-1469. doi: 10.2147/JIR.S402290. eCollection 2023. J Inflamm Res. 2023. PMID: 37042016 Free PMC article. Review.
-
Shaping Up Mitochondria in Diabetic Nephropathy.Kidney360. 2020 Sep 24;1(9):982-992. doi: 10.34067/kid.0002352020. Kidney360. 2020. PMID: 34189465 Free PMC article. Review.
-
Roles of Mitochondrial DNA Damage in Kidney Diseases: A New Biomarker.Int J Mol Sci. 2022 Dec 2;23(23):15166. doi: 10.3390/ijms232315166. Int J Mol Sci. 2022. PMID: 36499488 Free PMC article. Review.
-
Mitochondriopathy Manifesting as Inherited Tubulointerstitial Nephropathy Without Symptomatic Other Organ Involvement.Kidney Int Rep. 2021 Jun 12;6(9):2514-2518. doi: 10.1016/j.ekir.2021.05.042. eCollection 2021 Sep. Kidney Int Rep. 2021. PMID: 34514217 Free PMC article. No abstract available.
-
Exploring the impact and utility of genomic sequencing in established CKD.Clin Kidney J. 2024 Feb 21;17(3):sfae043. doi: 10.1093/ckj/sfae043. eCollection 2024 Mar. Clin Kidney J. 2024. PMID: 38464959 Free PMC article. Review.
References
-
- USRDS. U.S. Renal Data System, USRDS 2011 Annual Data Report: Atlas of End-Stage Renal Disease in the United States, National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases, Bethesda, MD, 2011. 2008 [2012]. Available from: http://www.usrds.org/adr_2008.htm.
-
- Eckardt KU, Alper SL, Antignac C, Bleyer AJ, Chauveau D, Dahan K, et al. Autosomal dominant tubulointerstitial kidney disease: diagnosis, classification, and management-A KDIGO consensus report. Kidney Int. 2015. Epub 2015/03/05. - PubMed
-
- Stewart JB, Chinnery PF. The dynamics of mitochondrial DNA heteroplasmy: implications for human health and disease. Nature Reviews Genetics. 2015. - PubMed
-
- Levine AP, Connor TM, Oygar DD, Neild GH, Segal AW, Maxwell PH, et al. Combinatorial Conflicting Homozygosity (CCH) analysis enables the rapid identification of shared genomic regions in the presence of multiple phenocopies. BMC genomics. 2015;16:163 Epub 2015/04/19. PubMed Central PMCID: PMC4364077. 10.1186/s12864-015-1360-4 - DOI - PMC - PubMed
-
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290(5806):457–65. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- MC_UP_1501/2/MRC_/Medical Research Council/United Kingdom
- MC_UU_12022/6/MRC_/Medical Research Council/United Kingdom
- 101876/Z/13/Z/WT_/Wellcome Trust/United Kingdom
- MR/K002201/2/MRC_/Medical Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- 100140/WT_/Wellcome Trust/United Kingdom
- G0701320/MRC_/Medical Research Council/United Kingdom
- G0802266/MRC_/Medical Research Council/United Kingdom
- 19710/WT_/Wellcome Trust/United Kingdom
- MR/K002201/1/MRC_/Medical Research Council/United Kingdom
- G1002528/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

