Refining the role of de novo protein-truncating variants in neurodevelopmental disorders by using population reference samples
- PMID: 28191890
- PMCID: PMC5496244
- DOI: 10.1038/ng.3789
Refining the role of de novo protein-truncating variants in neurodevelopmental disorders by using population reference samples
Abstract
Recent research has uncovered an important role for de novo variation in neurodevelopmental disorders. Using aggregated data from 9,246 families with autism spectrum disorder, intellectual disability, or developmental delay, we found that ∼1/3 of de novo variants are independently present as standing variation in the Exome Aggregation Consortium's cohort of 60,706 adults, and these de novo variants do not contribute to neurodevelopmental risk. We further used a loss-of-function (LoF)-intolerance metric, pLI, to identify a subset of LoF-intolerant genes containing the observed signal of associated de novo protein-truncating variants (PTVs) in neurodevelopmental disorders. LoF-intolerant genes also carry a modest excess of inherited PTVs, although the strongest de novo-affected genes contribute little to this excess, thus suggesting that the excess of inherited risk resides in lower-penetrant genes. These findings illustrate the importance of population-based reference cohorts for the interpretation of candidate pathogenic variants, even for analyses of complex diseases and de novo variation.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

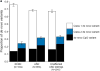


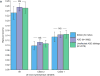





Similar articles
-
Diagnostic Yield and Novel Candidate Genes by Exome Sequencing in 152 Consanguineous Families With Neurodevelopmental Disorders.JAMA Psychiatry. 2017 Mar 1;74(3):293-299. doi: 10.1001/jamapsychiatry.2016.3798. JAMA Psychiatry. 2017. PMID: 28097321
-
De novo variants in neurodevelopmental disorders-experiences from a tertiary care center.Clin Genet. 2021 Jul;100(1):14-28. doi: 10.1111/cge.13946. Epub 2021 Mar 1. Clin Genet. 2021. PMID: 33619735
-
De novo variants in neurodevelopmental disorders with epilepsy.Nat Genet. 2018 Jul;50(7):1048-1053. doi: 10.1038/s41588-018-0143-7. Epub 2018 Jun 25. Nat Genet. 2018. PMID: 29942082
-
Estimating contribution of rare non-coding variants to neuropsychiatric disorders.Psychiatry Clin Neurosci. 2019 Jan;73(1):2-10. doi: 10.1111/pcn.12774. Epub 2018 Oct 6. Psychiatry Clin Neurosci. 2019. PMID: 30293238 Review.
-
Human GRIN2B variants in neurodevelopmental disorders.J Pharmacol Sci. 2016 Oct;132(2):115-121. doi: 10.1016/j.jphs.2016.10.002. Epub 2016 Oct 19. J Pharmacol Sci. 2016. PMID: 27818011 Free PMC article. Review.
Cited by
-
Deleterious ZNRF3 germline variants cause neurodevelopmental disorders with mirror brain phenotypes via domain-specific effects on Wnt/β-catenin signaling.Am J Hum Genet. 2024 Sep 5;111(9):1994-2011. doi: 10.1016/j.ajhg.2024.07.016. Epub 2024 Aug 20. Am J Hum Genet. 2024. PMID: 39168120 Free PMC article.
-
Novel variant in the CNNM2 gene associated with dominant hypomagnesemia.PLoS One. 2020 Sep 30;15(9):e0239965. doi: 10.1371/journal.pone.0239965. eCollection 2020. PLoS One. 2020. PMID: 32997713 Free PMC article.
-
The role of SLC12A family of cation-chloride cotransporters and drug discovery methodologies.J Pharm Anal. 2023 Dec;13(12):1471-1495. doi: 10.1016/j.jpha.2023.09.002. Epub 2023 Sep 9. J Pharm Anal. 2023. PMID: 38223443 Free PMC article. Review.
-
Combining exome/genome sequencing with data repository analysis reveals novel gene-disease associations for a wide range of genetic disorders.Genet Med. 2021 Aug;23(8):1551-1568. doi: 10.1038/s41436-021-01159-0. Epub 2021 Apr 19. Genet Med. 2021. PMID: 33875846 Free PMC article.
-
Loss-of-function variants in MYCBP2 cause neurobehavioural phenotypes and corpus callosum defects.Brain. 2023 Apr 19;146(4):1373-1387. doi: 10.1093/brain/awac364. Brain. 2023. PMID: 36200388 Free PMC article.
References
-
- Developmental D.M.N.S.Y. & Investigators P. Morbidity mortality weekly report. Surveillance summaries. Vol. 63. Washington, DC: 2014. Prevalence of autism spectrum disorder among children aged 8 years-autism and developmental disabilities monitoring network 11 sites, United States, 2010; p. 1. 2002. - PubMed
MeSH terms
Grants and funding
- K01 MH099286/MH/NIMH NIH HHS/United States
- R01 MH097849/MH/NIMH NIH HHS/United States
- U01 MH100229/MH/NIMH NIH HHS/United States
- U01 MH111660/MH/NIMH NIH HHS/United States
- R56 MH097849/MH/NIMH NIH HHS/United States
- U01 MH100209/MH/NIMH NIH HHS/United States
- T32 HG002295/HG/NHGRI NIH HHS/United States
- U01 MH100239/MH/NIMH NIH HHS/United States
- U01 MH111658/MH/NIMH NIH HHS/United States
- U01 MH111661/MH/NIMH NIH HHS/United States
- U54 DK105566/DK/NIDDK NIH HHS/United States
- R01 GM104371/GM/NIGMS NIH HHS/United States
- U01 MH100233/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

