Highly Signal-Responsive Gene Regulatory Network Governing Myxococcus Development
- PMID: 27916428
- PMCID: PMC5182100
- DOI: 10.1016/j.tig.2016.10.006
Highly Signal-Responsive Gene Regulatory Network Governing Myxococcus Development
Abstract
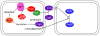
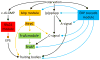
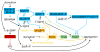
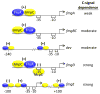
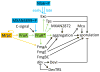
The bacterium Myxococcus xanthus undergoes multicellular development when starved. Thousands of cells build mounds in which some differentiate into spores. This remarkable feat and the genetic tractability of Myxococcus provide a unique opportunity to understand the evolution of gene regulatory networks (GRNs). Recent work has revealed a GRN involving interconnected cascades of signal-responsive transcriptional activators. Initially, starvation-induced intracellular signals direct changes in gene expression. Subsequently, self-generated extracellular signals provide morphological cues that regulate certain transcriptional activators. However, signals for many of the activators remain to be discovered. A key insight is that activators often work combinatorially, allowing signal integration. The Myxococcus GRN differs strikingly from those governing sporulation of Bacillus and Streptomyces, suggesting that Myxococcus evolved a highly signal-responsive GRN to enable complex multicellular development.
Keywords: Myxococcus xanthus; bacterial development; gene regulatory network; signal transduction; sporulation.
Copyright © 2016 Elsevier Ltd. All rights reserved.
Figures

Similar articles
-
The dev Operon Regulates the Timing of Sporulation during Myxococcus xanthus Development.J Bacteriol. 2017 Apr 25;199(10):e00788-16. doi: 10.1128/JB.00788-16. Print 2017 May 15. J Bacteriol. 2017. PMID: 28264995 Free PMC article.
-
The enhancer binding protein Nla6 regulates developmental genes that are important for Myxococcus xanthus sporulation.J Bacteriol. 2015 Apr;197(7):1276-87. doi: 10.1128/JB.02408-14. Epub 2015 Feb 2. J Bacteriol. 2015. PMID: 25645554 Free PMC article.
-
devI is an evolutionarily young negative regulator of Myxococcus xanthus development.J Bacteriol. 2015 Apr;197(7):1249-62. doi: 10.1128/JB.02542-14. Epub 2015 Feb 2. J Bacteriol. 2015. PMID: 25645563 Free PMC article.
-
Two-Component Signal Transduction Systems That Regulate the Temporal and Spatial Expression of Myxococcus xanthus Sporulation Genes.J Bacteriol. 2015 Sep 14;198(3):377-85. doi: 10.1128/JB.00474-15. Print 2016 Feb 1. J Bacteriol. 2015. PMID: 26369581 Free PMC article. Review.
-
Regulations governing the multicellular lifestyle of Myxococcus xanthus.Curr Opin Microbiol. 2016 Dec;34:104-110. doi: 10.1016/j.mib.2016.08.009. Epub 2016 Sep 17. Curr Opin Microbiol. 2016. PMID: 27648756 Review.
Cited by
-
Cell density, alignment, and orientation correlate with C-signal-dependent gene expression during Myxococcus xanthus development.Proc Natl Acad Sci U S A. 2021 Nov 9;118(45):e2111706118. doi: 10.1073/pnas.2111706118. Proc Natl Acad Sci U S A. 2021. PMID: 34732578 Free PMC article.
-
Complete Genome Sequence of the Fruiting Myxobacterium Myxococcus macrosporus Strain DSM 14697, Generated by PacBio Sequencing.Genome Announc. 2017 Oct 5;5(40):e01127-17. doi: 10.1128/genomeA.01127-17. Genome Announc. 2017. PMID: 28983009 Free PMC article.
-
Complete Genome Sequence of the Fruiting Myxobacterium Melittangium boletus DSM 14713.Genome Announc. 2017 Nov 9;5(45):e01262-17. doi: 10.1128/genomeA.01262-17. Genome Announc. 2017. PMID: 29122879 Free PMC article.
-
An ambruticin-sensing complex modulates Myxococcus xanthus development and mediates myxobacterial interspecies communication.Nat Commun. 2020 Nov 4;11(1):5563. doi: 10.1038/s41467-020-19384-7. Nat Commun. 2020. PMID: 33149152 Free PMC article.
-
CRP-Like Transcriptional Regulator MrpC Curbs c-di-GMP and 3',3'-cGAMP Nucleotide Levels during Development in Myxococcus xanthus.mBio. 2021 Feb 22;13(1):e0004422. doi: 10.1128/mbio.00044-22. Epub 2022 Feb 15. mBio. 2021. PMID: 35164555 Free PMC article.
References
-
- Yang Z, Higgs P, editors. Myxobacteria: genomics, cellular and molecular biology. Caister Academic Press; Norfolk, UK: 2014.
-
- Rajagopalan R, et al. Developmental gene regulation. In: Yang Z, Higgs P, editors. Myxobacteria: genomics, cellular and molecular biology. Caister Academic Press; Norfolk, UK: 2014. pp. 105–126.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

