Structure of a peptide:N-glycanase-Rad23 complex: insight into the deglycosylation for denatured glycoproteins
- PMID: 15964983
- PMCID: PMC1166607
- DOI: 10.1073/pnas.0502082102
Structure of a peptide:N-glycanase-Rad23 complex: insight into the deglycosylation for denatured glycoproteins
Abstract
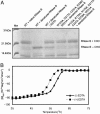
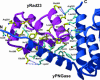
In eukaryotes, misfolded proteins must be distinguished from correctly folded proteins during folding and transport processes by quality control systems. Yeast peptide:N-glycanase (yPNGase) specifically deglycosylates the denatured form of N-linked glycoproteins in the cytoplasm and assists proteasome-mediated glycoprotein degradation by forming a complex with 26S proteasome through DNA repair protein, yRad23. Here, we describe the crystal structures of a yPNGase and XPC-binding domain of yRad23 (yRad23XBD, residues 238-309) complex and of a yPNGase-yRad23XBD complex bound to a caspase inhibitor, Z-VAD-fmk. yPNGase is formed with three domains, a core domain containing a Cys-His-Asp triad, a Zn-binding domain, and a Rad23-binding domain. Both N- and C-terminal helices of yPNGase interact with yRad23 through extensive hydrophobic interactions. The active site of yPNGase is located in a deep cleft that is formed with residues conserved in all PNGase members, and three sugar molecules are bound to this cleft. Complex structures in conjunction with mutational analyses revealed that the walls of the cleft block access to the active site of yPNGase by native glycoprotein, whereas the cleft is sufficiently wide to accommodate denatured glycoprotein, thus explaining the specificity of PNGase for denatured substrates.
Figures
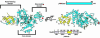



Similar articles
-
The N-terminus of yeast peptide: N-glycanase interacts with the DNA repair protein Rad23.Biochem Biophys Res Commun. 2004 Oct 8;323(1):149-55. doi: 10.1016/j.bbrc.2004.08.061. Biochem Biophys Res Commun. 2004. PMID: 15351714
-
Structural and mutational studies on the importance of oligosaccharide binding for the activity of yeast PNGase.Glycobiology. 2009 Feb;19(2):118-25. doi: 10.1093/glycob/cwn108. Epub 2008 Oct 14. Glycobiology. 2009. PMID: 18854368 Free PMC article.
-
Fluorescently labeled inhibitor for profiling cytoplasmic peptide:N-glycanase.Glycobiology. 2007 Oct;17(10):1070-6. doi: 10.1093/glycob/cwm079. Epub 2007 Jul 19. Glycobiology. 2007. PMID: 17640972
-
Physiological and molecular functions of the cytosolic peptide:N-glycanase.Semin Cell Dev Biol. 2015 May;41:110-20. doi: 10.1016/j.semcdb.2014.11.009. Epub 2014 Dec 2. Semin Cell Dev Biol. 2015. PMID: 25475175 Review.
-
Cytoplasmic peptide:N-glycanase (PNGase) in eukaryotic cells: occurrence, primary structure, and potential functions.FASEB J. 2002 May;16(7):635-41. doi: 10.1096/fj.01-0889rev. FASEB J. 2002. PMID: 11978727 Review.
Cited by
-
Structure of the catalytic domain of the Salmonella virulence factor SseI.Acta Crystallogr D Biol Crystallogr. 2012 Dec;68(Pt 12):1613-21. doi: 10.1107/S0907444912039042. Epub 2012 Nov 9. Acta Crystallogr D Biol Crystallogr. 2012. PMID: 23151626 Free PMC article.
-
Multiple modes of interaction of the deglycosylation enzyme, mouse peptide N-glycanase, with the proteasome.Proc Natl Acad Sci U S A. 2005 Nov 1;102(44):15809-14. doi: 10.1073/pnas.0507155102. Epub 2005 Oct 25. Proc Natl Acad Sci U S A. 2005. PMID: 16249333 Free PMC article.
-
Structural and biochemical studies of the C-terminal domain of mouse peptide-N-glycanase identify it as a mannose-binding module.Proc Natl Acad Sci U S A. 2006 Nov 14;103(46):17214-9. doi: 10.1073/pnas.0602954103. Epub 2006 Nov 6. Proc Natl Acad Sci U S A. 2006. PMID: 17088551 Free PMC article.
-
Human DNA-Damage-Inducible 2 Protein Is Structurally and Functionally Distinct from Its Yeast Ortholog.Sci Rep. 2016 Jul 27;6:30443. doi: 10.1038/srep30443. Sci Rep. 2016. PMID: 27461074 Free PMC article.
-
Comprehensive Analysis of the Structure and Function of Peptide:N-Glycanase 1 and Relationship with Congenital Disorder of Deglycosylation.Nutrients. 2022 Apr 19;14(9):1690. doi: 10.3390/nu14091690. Nutrients. 2022. PMID: 35565658 Free PMC article. Review.
References
-
- Helenius, A. & Aebi, M. (2001) Science 291, 2364-2369. - PubMed
-
- Goldberg, A. L. (2003) Nature 426, 895-899. - PubMed
-
- Rutishauser, J. & Spiess, M. (2002) Swiss Med. Wkly. 132, 211-222. - PubMed
-
- Bailey, C. K., Andriola, I. F., Kampinga, H. H. & Merry, D. E. (2002) Hum. Mol. Genet. 11, 515-523. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

