Identification and characterization of the human ARD1-NATH protein acetyltransferase complex
- PMID: 15496142
- PMCID: PMC1134861
- DOI: 10.1042/BJ20041071
Identification and characterization of the human ARD1-NATH protein acetyltransferase complex
Abstract
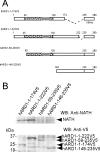
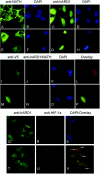
Protein acetyltransferases and deacetylases have been implicated in oncogenesis, apoptosis and cell cycle regulation. Most of the protein acetyltransferases described acetylate epsilon-amino groups of lysine residues within proteins. Mouse ARD1 (homologue of yeast Ard1p, where Ard1p stands for arrest defective 1 protein) is the only known protein acetyltransferase catalysing acetylation of proteins at both alpha-(N-terminus) and epsilon-amino groups. Yeast Ard1p interacts with Nat1p (N-acetyltransferase 1 protein) to form a functional NAT (N-acetyltransferase). We now describe the human homologue of Nat1p, NATH (NAT human), as the partner of the hARD1 (human ARD1) protein. Included in the characterization of the NATH and hARD1 proteins is the following: (i) endogenous NATH and hARD1 proteins are expressed in human epithelial, glioma and promyelocytic cell lines; (ii) NATH and hARD1 form a stable complex, as investigated by reciprocal immunoprecipitations followed by MS analysis; (iii) NATH-hARD1 complex expresses N-terminal acetylation activity; (iv) NATH and hARD1 interact with ribosomal subunits, indicating a co-translational acetyltransferase function; (v) NATH is localized in the cytoplasm, whereas hARD1 localizes both to the cytoplasm and nucleus; (vi) hARD1 partially co-localizes in nuclear spots with the transcription factor HIF-1alpha (hypoxia-inducible factor 1alpha), a known epsilon-amino substrate of ARD1; (vii) NATH and hARD1 are cleaved during apoptosis, resulting in a decreased NAT activity. This study identifies the human homologues of the yeast Ard1p and Nat1p proteins and presents new aspects of the NATH and hARD1 proteins relative to their yeast homologues.
Figures


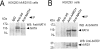


Similar articles
-
Characterization of hARD2, a processed hARD1 gene duplicate, encoding a human protein N-alpha-acetyltransferase.BMC Biochem. 2006 Apr 25;7:13. doi: 10.1186/1471-2091-7-13. BMC Biochem. 2006. PMID: 16638120 Free PMC article.
-
Expression of N-acetyl transferase human and human Arrest defective 1 proteins in thyroid neoplasms.Thyroid. 2005 Oct;15(10):1131-6. doi: 10.1089/thy.2005.15.1131. Thyroid. 2005. PMID: 16279846
-
Induction of apoptosis in human cells by RNAi-mediated knockdown of hARD1 and NATH, components of the protein N-alpha-acetyltransferase complex.Oncogene. 2006 Jul 20;25(31):4350-60. doi: 10.1038/sj.onc.1209469. Epub 2006 Mar 6. Oncogene. 2006. PMID: 16518407
-
The protein acetyltransferase ARD1: a novel cancer drug target?Curr Cancer Drug Targets. 2008 Nov;8(7):545-53. doi: 10.2174/156800908786241113. Curr Cancer Drug Targets. 2008. PMID: 18991565 Review.
-
Versatility of ARD1/NAA10-mediated protein lysine acetylation.Exp Mol Med. 2018 Jul 27;50(7):1-13. doi: 10.1038/s12276-018-0100-7. Exp Mol Med. 2018. PMID: 30054464 Free PMC article. Review.
Cited by
-
Synthesis of Recombinant Human Hemoglobin With NH2 -Terminal Acetylation in Escherichia coli.Curr Protoc Protein Sci. 2020 Sep;101(1):e112. doi: 10.1002/cpps.112. Curr Protoc Protein Sci. 2020. PMID: 32687676 Free PMC article.
-
Protein N-Terminal Acetylation: Structural Basis, Mechanism, Versatility, and Regulation.Trends Biochem Sci. 2021 Jan;46(1):15-27. doi: 10.1016/j.tibs.2020.08.005. Epub 2020 Sep 8. Trends Biochem Sci. 2021. PMID: 32912665 Free PMC article. Review.
-
A repository of Ogden syndrome patient derived iPSC lines and isogenic pairs by X-chromosome screening and genome-editing.bioRxiv [Preprint]. 2024 Sep 28:2024.09.28.615067. doi: 10.1101/2024.09.28.615067. bioRxiv. 2024. PMID: 39386428 Free PMC article. Preprint.
-
TSG101 associates with PARP1 and is essential for PARylation and DNA damage-induced NF-κB activation.EMBO J. 2022 Nov 2;41(21):e110372. doi: 10.15252/embj.2021110372. Epub 2022 Sep 20. EMBO J. 2022. PMID: 36124865 Free PMC article.
-
A Case of NAA10-related Syndrome With Prolonged QTc Treated With a Subcutaneous Implantable Cardioverter Defibrillator After Ventricular Fibrillation.CJC Pediatr Congenit Heart Dis. 2022 Oct 7;1(6):270-273. doi: 10.1016/j.cjcpc.2022.10.001. eCollection 2022 Dec. CJC Pediatr Congenit Heart Dis. 2022. PMID: 37969489 Free PMC article.
References
-
- Cohen H. Y., Lavu S., Bitterman K. J., Hekking B., Imahiyerobo T. A., Miller C., Frye R., Ploegh H., Kessler B. M., Sinclair D. A. Acetylation of the C terminus of Ku70 by CBP and PCAF controls Bax-mediated apoptosis. Mol. Cell. 2004;13:627–638. - PubMed
-
- Fu M., Wang C., Wang J., Zafonte B. T., Lisanti M. P., Pestell R. G. Acetylation in hormone signaling and the cell cycle. Cytokine Growth Factor Rev. 2002;13:259–276. - PubMed
-
- Di Gennaro E., Bruzzese F., Caraglia M., Abruzzese A., Budillon A. Acetylation of proteins as novel target for antitumor therapy: review article. Amino Acids. 2004;26:435–441. - PubMed
-
- Sugiura N., Adams S. M., Corriveau R. A. An evolutionarily conserved N-terminal acetyltransferase complex associated with neuronal development. J. Biol. Chem. 2003;278:40113–40120. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

