The ribosomal translocase homologue Snu114p is involved in unwinding U4/U6 RNA during activation of the spliceosome
- PMID: 12189173
- PMCID: PMC1084225
- DOI: 10.1093/embo-reports/kvf172
The ribosomal translocase homologue Snu114p is involved in unwinding U4/U6 RNA during activation of the spliceosome
Abstract
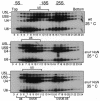
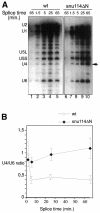
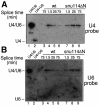
Snu114p is a yeast U5 snRNP protein homologous to the ribosomal elongation factor EF-2. Snu114p exhibits the same domain structure as EF-2, including the G-domain, but with an additional N-terminal domain. To test whether Snu114p in the spliceosome is involved in rearranging RNA secondary structures (by analogy to EF-2 in the ribosome), we created conditionally lethal mutants. Deletion of this N-terminal domain (snu114 Delta N) leads to a temperature-sensitive phenotype at 37 degrees C and a pre-mRNA splicing defect in vivo. Heat treatment of snu114 Delta N extracts blocked splicing in vitro before the first step. The snu114 Delta N still associates with the tri-snRNP, and the stability of this particle is not significantly impaired by thermal inactivation. Heat treatment of snu114 Delta N extracts resulted in accumulation of arrested spliceosomes in which the U4 RNA was not efficiently released, and we show that U4 is still base paired with the U6 RNA. This suggests that Snu114p is involved, directly or indirectly, in the U4/U6 unwinding, an essential step towards spliceosome activation.
Figures


Similar articles
-
The EF-G-like GTPase Snu114p regulates spliceosome dynamics mediated by Brr2p, a DExD/H box ATPase.Mol Cell. 2006 Aug 4;23(3):389-99. doi: 10.1016/j.molcel.2006.05.043. Mol Cell. 2006. PMID: 16885028 Free PMC article.
-
Analysis of synthetic lethality reveals genetic interactions between the GTPase Snu114p and snRNAs in the catalytic core of the Saccharomyces cerevisiae spliceosome.Genetics. 2009 Oct;183(2):497-515-1SI-4SI. doi: 10.1534/genetics.109.107243. Epub 2009 Jul 20. Genetics. 2009. PMID: 19620389 Free PMC article.
-
Mutagenesis suggests several roles of Snu114p in pre-mRNA splicing.J Biol Chem. 2003 Jul 25;278(30):28324-34. doi: 10.1074/jbc.M303043200. Epub 2003 May 7. J Biol Chem. 2003. PMID: 12736260
-
Assembly of Snu114 into U5 snRNP requires Prp8 and a functional GTPase domain.RNA. 2006 May;12(5):862-71. doi: 10.1261/rna.2319806. Epub 2006 Mar 15. RNA. 2006. PMID: 16540695 Free PMC article.
-
The role of Snu114p during pre-mRNA splicing.Biochem Soc Trans. 2008 Jun;36(Pt 3):551-3. doi: 10.1042/BST0360551. Biochem Soc Trans. 2008. PMID: 18482006 Review.
Cited by
-
Acetylation by the transcriptional coactivator Gcn5 plays a novel role in co-transcriptional spliceosome assembly.PLoS Genet. 2009 Oct;5(10):e1000682. doi: 10.1371/journal.pgen.1000682. Epub 2009 Oct 16. PLoS Genet. 2009. PMID: 19834536 Free PMC article.
-
The EF-G-like GTPase Snu114p regulates spliceosome dynamics mediated by Brr2p, a DExD/H box ATPase.Mol Cell. 2006 Aug 4;23(3):389-99. doi: 10.1016/j.molcel.2006.05.043. Mol Cell. 2006. PMID: 16885028 Free PMC article.
-
Novel regulatory principles of the spliceosomal Brr2 RNA helicase and links to retinal disease in humans.RNA Biol. 2014;11(4):298-312. doi: 10.4161/rna.28353. Epub 2014 Mar 5. RNA Biol. 2014. PMID: 24643059 Free PMC article. Review.
-
Spliceosome structure and function.Cold Spring Harb Perspect Biol. 2011 Jul 1;3(7):a003707. doi: 10.1101/cshperspect.a003707. Cold Spring Harb Perspect Biol. 2011. PMID: 21441581 Free PMC article. Review.
-
Analysis of synthetic lethality reveals genetic interactions between the GTPase Snu114p and snRNAs in the catalytic core of the Saccharomyces cerevisiae spliceosome.Genetics. 2009 Oct;183(2):497-515-1SI-4SI. doi: 10.1534/genetics.109.107243. Epub 2009 Jul 20. Genetics. 2009. PMID: 19620389 Free PMC article.
References
-
- Achsel T., Ahrens, K., Brahms, H., Teigelkamp, S. and Lührmann, R. (1998) The human U5-220kD protein (hPrp8) forms a stable RNA-free complex with several U5-specific proteins, including an RNA unwindase, a homologue of ribosomal elongation factor EF-2, and a novel WD-40 protein. Mol. Cell. Biol., 18, 6756–6766. - PMC - PubMed
-
- Burge C.B., Tuschl, T. and Sharp, P.A. (1999) Splicing of precursors to mRNAs by the spliceosomes. In Gesteland, R.F. and Atkins, J.F. (eds), The RNA World. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 525–560.
-
- Chen J.Y., Stands, L., Staley, J.P., Jackups, R.R., Jr, Latus, L.J. and Chang, T.H. (2001) Specific alterations of U1-C protein or U1 small nuclear RNA can eliminate the requirement of Prp28p, an essential DEAD box splicing factor. Mol. Cell, 7, 227–232. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

