
Han-Yu Shih, Ph.D., Earl Stadtman Investigator and chief of the NEI Neuro-Immune Regulome Unit.
According to a new study from the National Eye Institute (NEI), the configuration of DNA loops in developing immune cells dictates how effectively the cells fight infection. The loops tether specific DNA regulatory regions to genes encoding key pro- and anti-inflammatory signaling molecules called cytokines. According to the new study, this arrangement determines cytokine expression level, which is essential for protection against pathogens. The study published in Immunity. NEI is part of the National Institutes of Health.
An effective immune system attacks pathogens while avoiding improper responses like allergies or autoimmune conditions. T cells are immune cells that recognize and distinguish foreign substances from the body’s own tissues and coordinate the body’s defenses. As T cells mature, they are primed by other immune cells to produce cytokines that help fight off pathogens, like viruses or bacteria. Dysregulation of these cytokines can lead to autoimmune disorders where the immune system attacks the body.
Turning on cytokine genes requires arrays of DNA elements called enhancers, which enable the cell to begin churning out active cytokine proteins. While many enhancers are close by the gene they drive, others are further away. DNA looping brings far-away enhancers into close proximity with their corresponding gene.
Han-Yu Shih, Ph.D., chief of the NEI Neuro-immune Regulome Unit, and colleagues found that a region of DNA – just 17 base pairs long – helps generate a DNA loop that regulates the expression of the pro-inflammatory cytokine IFN-γ, enabling normal development of T cells. Mice missing this short stretch of non-coding DNA were less able to clear infections by the common parasite Toxoplasma gondii—and are more likely to die from the infection—than normal mice. The researchers observed that in mice without the non-coding regulatory DNA region, Th1 cells, a type of T cell, produced less IFN-γ protein, resulting in an ineffective immune response. NK cells, another type of immune cell that produces IFN-γ, were unaffected by loss of the regulatory region, suggesting that this mechanism is specific to T cells.
“This study affirms the significance of non-coding DNA regions in regulating the immune system; harnessing this information may help us one day combat infectious, autoimmune and inflammatory diseases of the brain and eyes,” said Shih.
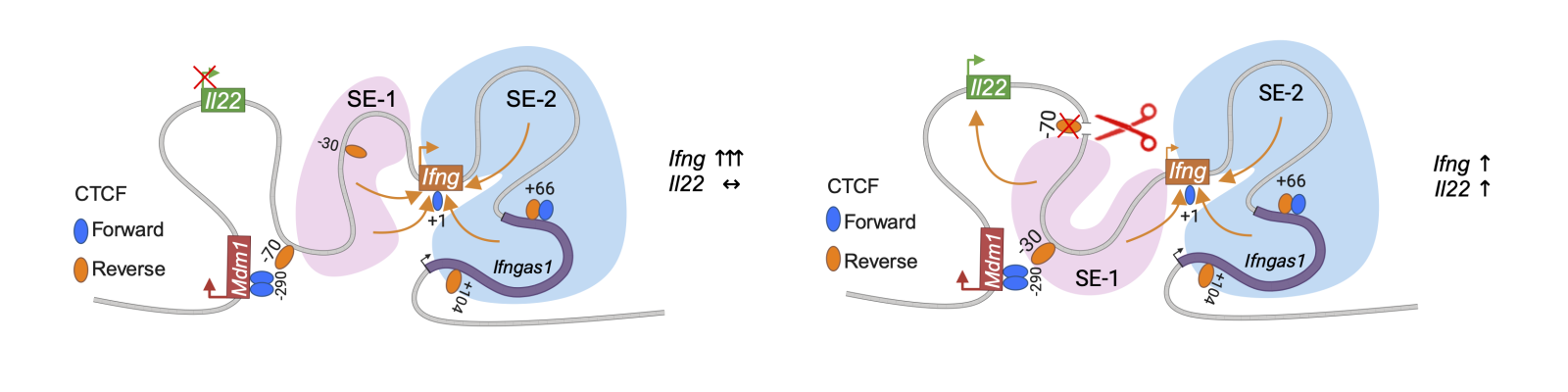
With 17bp regulatory region present, DNA looping induces robust IFN-γ expression. Without the regulatory region, DNA loops are altered, reducing IFN-γ expression. Image credit: Han-Yu Shih, Ph.D.
Reference: Liu C, Nagashima H, et al. A CTCF-binding site in the Mdm1-Il22-Ifng locus shapes cytokine expression profiles and plays a critical role in early Th1 cell fate specification. Immunity. 2024 May 14;57(5):1005-1018.e7. doi: 10.1016/j.immuni.2024.04.007.
This study was primarily funded by the NEI Intramural Research Program. Intramural Research Programs of the National Institute of Arthritis and Musculoskeletal and Skin Diseases and the National Institute of Allergy and Infectious Diseases also supported the research.