Gene copy-number polymorphism caused by retrotransposition in humans
- PMID: 23359205
- PMCID: PMC3554589
- DOI: 10.1371/journal.pgen.1003242
Gene copy-number polymorphism caused by retrotransposition in humans
Abstract
The era of whole-genome sequencing has revealed that gene copy-number changes caused by duplication and deletion events have important evolutionary, functional, and phenotypic consequences. Recent studies have therefore focused on revealing the extent of variation in copy-number within natural populations of humans and other species. These studies have found a large number of copy-number variants (CNVs) in humans, many of which have been shown to have clinical or evolutionary importance. For the most part, these studies have failed to detect an important class of gene copy-number polymorphism: gene duplications caused by retrotransposition, which result in a new intron-less copy of the parental gene being inserted into a random location in the genome. Here we describe a computational approach leveraging next-generation sequence data to detect gene copy-number variants caused by retrotransposition (retroCNVs), and we report the first genome-wide analysis of these variants in humans. We find that retroCNVs account for a substantial fraction of gene copy-number differences between any two individuals. Moreover, we show that these variants may often result in expressed chimeric transcripts, underscoring their potential for the evolution of novel gene functions. By locating the insertion sites of these duplicates, we are able to show that retroCNVs have had an important role in recent human adaptation, and we also uncover evidence that positive selection may currently be driving multiple retroCNVs toward fixation. Together these findings imply that retroCNVs are an especially important class of polymorphism, and that future studies of copy-number variation should search for these variants in order to illuminate their potential evolutionary and functional relevance.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
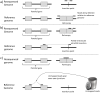
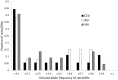
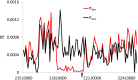
Similar articles
-
An evolving view of copy number variants.Curr Genet. 2019 Dec;65(6):1287-1295. doi: 10.1007/s00294-019-00980-0. Epub 2019 May 10. Curr Genet. 2019. PMID: 31076843 Review.
-
An Evolutionary Perspective on the Impact of Genomic Copy Number Variation on Human Health.J Mol Evol. 2020 Jan;88(1):104-119. doi: 10.1007/s00239-019-09911-6. Epub 2019 Sep 14. J Mol Evol. 2020. PMID: 31522275 Review.
-
Next-generation sequencing of duplication CNVs reveals that most are tandem and some create fusion genes at breakpoints.Am J Hum Genet. 2015 Feb 5;96(2):208-20. doi: 10.1016/j.ajhg.2014.12.017. Epub 2015 Jan 29. Am J Hum Genet. 2015. PMID: 25640679 Free PMC article.
-
A Highly Specific Genome-Wide Association Study Integrated with Transcriptome Data Reveals the Contribution of Copy Number Variations to Specialized Metabolites in Arabidopsis thaliana Accessions.Mol Biol Evol. 2017 Dec 1;34(12):3111-3122. doi: 10.1093/molbev/msx234. Mol Biol Evol. 2017. PMID: 28961930
-
A computational framework discovers new copy number variants with functional importance.PLoS One. 2011 Mar 29;6(3):e17539. doi: 10.1371/journal.pone.0017539. PLoS One. 2011. PMID: 21479260 Free PMC article.
Cited by
-
Adaptive potential of genomic structural variation in human and mammalian evolution.Brief Funct Genomics. 2015 Sep;14(5):358-68. doi: 10.1093/bfgp/elv019. Epub 2015 May 23. Brief Funct Genomics. 2015. PMID: 26003631 Free PMC article. Review.
-
The Influence of LINE-1 and SINE Retrotransposons on Mammalian Genomes.Microbiol Spectr. 2015 Apr;3(2):MDNA3-0061-2014. doi: 10.1128/microbiolspec.MDNA3-0061-2014. Microbiol Spectr. 2015. PMID: 26104698 Free PMC article. Review.
-
A Genome-Wide Landscape of Retrocopies in Primate Genomes.Genome Biol Evol. 2015 Jul 29;7(8):2265-75. doi: 10.1093/gbe/evv142. Genome Biol Evol. 2015. PMID: 26224704 Free PMC article.
-
Mapping and characterization of structural variation in 17,795 human genomes.Nature. 2020 Jul;583(7814):83-89. doi: 10.1038/s41586-020-2371-0. Epub 2020 May 27. Nature. 2020. PMID: 32460305 Free PMC article.
-
An evolving view of copy number variants.Curr Genet. 2019 Dec;65(6):1287-1295. doi: 10.1007/s00294-019-00980-0. Epub 2019 May 10. Curr Genet. 2019. PMID: 31076843 Review.
References
-
- Demuth JP, De Bie T, Stajich JE, Cristianini N, Hahn MW (2006) The evolution of mammalian gene families. PLoS ONE 1: e85 doi:10.1371/journal.pone.0000085. - DOI - PMC - PubMed
-
- Greenberg AJ, Moran JR, Fang S, Wu CI (2006) Adaptive loss of an old duplicated gene during incipient speciation. Mol Biol Evol 23: 401–410. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

