Genomic drift and copy number variation of sensory receptor genes in humans
- PMID: 18077390
- PMCID: PMC2154446
- DOI: 10.1073/pnas.0709956104
Genomic drift and copy number variation of sensory receptor genes in humans
Abstract
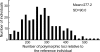
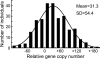
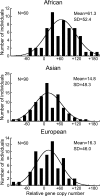
The number of sensory receptor genes varies extensively among different mammalian species. This variation is believed to be caused partly by physiological requirements of animals and partly by genomic drift due to random duplication and deletion of genes. If the contribution of genomic drift is substantial, each species should contain a significant amount of copy number variation (CNV). We therefore investigated CNVs in sensory receptor genes among 270 healthy humans by using published CNV data. The results indicated that olfactory receptor (OR), taste receptor type 2, and vomeronasal receptor type 1 genes show a high level of intraspecific CNVs. In particular, >30% of the approximately 800 OR gene loci in humans were polymorphic with respect to copy number, and two randomly chosen individuals showed a copy number difference of approximately 11 in functional OR genes on average. There was no significant difference in the amount of CNVs between functional and nonfunctional OR genes. Because pseudogenes are expected to evolve in a neutral fashion, this observation suggests that functional OR genes also have evolved in a similar manner with respect to copy number change. In addition, we found that the evolutionary change of copy number of OR genes approximately follows the Gaussian process in probability theory, and the copy number divergence between populations has increased with evolutionary time. We therefore conclude that genomic drift plays an important role for generating intra- and interspecific CNVs of sensory receptor genes. Similar results were obtained when all annotated genes were analyzed.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
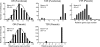

Comment in
-
The drifting human genome.Proc Natl Acad Sci U S A. 2007 Dec 18;104(51):20147-8. doi: 10.1073/pnas.0710524104. Proc Natl Acad Sci U S A. 2007. PMID: 18093959 Free PMC article. No abstract available.
Similar articles
-
Genomic drift and evolution of microsatellite DNAs in human populations.Mol Biol Evol. 2009 Aug;26(8):1835-40. doi: 10.1093/molbev/msp091. Epub 2009 Apr 30. Mol Biol Evol. 2009. PMID: 19406937
-
Genomic drift and copy number variation of chemosensory receptor genes in humans and mice.Cytogenet Genome Res. 2008;123(1-4):263-9. doi: 10.1159/000184716. Epub 2009 Mar 11. Cytogenet Genome Res. 2008. PMID: 19287163 Free PMC article.
-
High-resolution copy-number variation map reflects human olfactory receptor diversity and evolution.PLoS Genet. 2008 Nov;4(11):e1000249. doi: 10.1371/journal.pgen.1000249. Epub 2008 Nov 7. PLoS Genet. 2008. PMID: 18989455 Free PMC article.
-
Copy number variation in chemokine superfamily: the complex scene of CCL3L-CCL4L genes in health and disease.Clin Exp Immunol. 2010 Oct;162(1):41-52. doi: 10.1111/j.1365-2249.2010.04224.x. Epub 2010 Aug 19. Clin Exp Immunol. 2010. PMID: 20659124 Free PMC article. Review.
-
The evolution of animal chemosensory receptor gene repertoires: roles of chance and necessity.Nat Rev Genet. 2008 Dec;9(12):951-63. doi: 10.1038/nrg2480. Nat Rev Genet. 2008. PMID: 19002141 Review.
Cited by
-
Comparative genomics and evolution of the alpha-defensin multigene family in primates.Mol Biol Evol. 2010 Oct;27(10):2333-43. doi: 10.1093/molbev/msq118. Epub 2010 May 9. Mol Biol Evol. 2010. PMID: 20457584 Free PMC article.
-
The evolution of neurosensation provides opportunities and constraints for phenotypic plasticity.Sci Rep. 2022 Jul 13;12(1):11883. doi: 10.1038/s41598-022-15583-y. Sci Rep. 2022. PMID: 35831328 Free PMC article.
-
Rare structural variants found in attention-deficit hyperactivity disorder are preferentially associated with neurodevelopmental genes.Mol Psychiatry. 2010 Jun;15(6):637-46. doi: 10.1038/mp.2009.57. Epub 2009 Jun 23. Mol Psychiatry. 2010. PMID: 19546859 Free PMC article.
-
Genetics of canine olfaction and receptor diversity.Mamm Genome. 2012 Feb;23(1-2):132-43. doi: 10.1007/s00335-011-9371-1. Epub 2011 Nov 13. Mamm Genome. 2012. PMID: 22080304 Review.
-
Identification of Structural Variation in Chimpanzees Using Optical Mapping and Nanopore Sequencing.Genes (Basel). 2020 Mar 4;11(3):276. doi: 10.3390/genes11030276. Genes (Basel). 2020. PMID: 32143403 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

