A 2000-Year-Old Bacillus stercoris Strain Sheds Light on the Evolution of Cyclic Antimicrobial Lipopeptide Synthesis
- PMID: 38399742
- PMCID: PMC10893106
- DOI: 10.3390/microorganisms12020338
A 2000-Year-Old Bacillus stercoris Strain Sheds Light on the Evolution of Cyclic Antimicrobial Lipopeptide Synthesis
Abstract
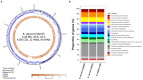
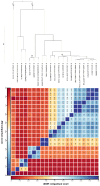
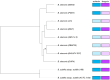
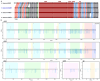
Some bacteria (notably the genera Bacillus and Clostridium) have the capacity to form endospores that can survive for millions of years in isolated habitats. The genomes of such ancient bacteria provide unique opportunities to understand bacterial evolution and metabolic capabilities over longer time scales. Herein, we sequenced the genome of a 2000-year-old bacterial strain (Mal05) isolated from intact apple seeds recovered during archaeological excavations of a Roman villa in Italy. Phylogenomic analyses revealed that this strain belongs to the species Bacillus stercoris and that it is placed in an early-branching position compared to most other strains of this species. Similar to other Bacillus species, B. stercoris Mal05 had been previously shown to possess antifungal activity. Its genome encodes all the genes necessary for the biosynthesis of fengycin and surfactin, two cyclic lipopeptides known to play a role in the competition of Bacilli with other microorganisms due to their antimicrobial activity. Comparative genomics and analyses of selective pressure demonstrate that these genes are present in all sequenced B. stercoris strains, despite the fact that they are not under strong purifying selection. Hence, these genes may not be essential for the fitness of these bacteria, but they can still provide a competitive advantage against other microorganisms present in the same environment.
Keywords: Bacillus stercoris; ancient bacteria; surfactin.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Promotion of Bacillus subtilis subsp. inaquosorum, Bacillus subtilis subsp. spizizenii and Bacillus subtilis subsp. stercoris to species status.Antonie Van Leeuwenhoek. 2020 Jan;113(1):1-12. doi: 10.1007/s10482-019-01354-9. Epub 2019 Nov 12. Antonie Van Leeuwenhoek. 2020. PMID: 31721032
-
Lipopeptide biodiversity in antifungal Bacillus strains isolated from Algeria.Arch Microbiol. 2018 Oct;200(8):1205-1216. doi: 10.1007/s00203-018-1537-8. Epub 2018 Jun 9. Arch Microbiol. 2018. PMID: 29947835
-
Biocontrol of Sclerotinia sclerotiorum (Lib.) de Bary on common bean by native lipopeptide-producer Bacillus strains.Microbiol Res. 2018 Jun;211:21-30. doi: 10.1016/j.micres.2018.04.003. Epub 2018 Apr 7. Microbiol Res. 2018. PMID: 29705203
-
Biological control of plant pathogens by Bacillus species.J Biotechnol. 2018 Nov 10;285:44-55. doi: 10.1016/j.jbiotec.2018.07.044. Epub 2018 Aug 30. J Biotechnol. 2018. PMID: 30172784 Review.
-
Key elements and regulation strategies of NRPSs for biosynthesis of lipopeptides by Bacillus.Appl Microbiol Biotechnol. 2020 Oct;104(19):8077-8087. doi: 10.1007/s00253-020-10801-x. Epub 2020 Aug 19. Appl Microbiol Biotechnol. 2020. PMID: 32813066 Review.
References
-
- Arriola L.A., Cooper A., Weyrich L.S. Palaeomicrobiology: Application of Ancient DNA Sequencing to Better Understand Bacterial Genome Evolution and Adaptation. Front. Ecol. Evol. 2020;8:40. doi: 10.3389/fevo.2020.00040. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

