Potential drug targets for asthma identified in the plasma and brain through Mendelian randomization analysis
- PMID: 37809092
- PMCID: PMC10551444
- DOI: 10.3389/fimmu.2023.1240517
Potential drug targets for asthma identified in the plasma and brain through Mendelian randomization analysis
Abstract
Background: Asthma is a heterogeneous disease, and the involvement of neurogenic inflammation is crucial in its development. The standardized treatments focus on alleviating symptoms. Despite the availability of medications for asthma, they have proven to be inadequate in controlling relapses and halting the progression of the disease. Therefore, there is a need for novel drug targets to prevent asthma.
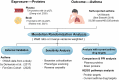
Methods: We utilized Mendelian randomization to investigate potential drug targets for asthma. We analyzed summary statistics from the UK Biobank and then replicated our findings in GWAS data by Demenais et al. and the FinnGen cohort. We obtained genetic instruments for 734 plasma and 73 brain proteins from recently reported GWAS. Next, we utilized reverse causal relationship analysis, Bayesian co-localization, and phenotype scanning as part of our sensitivity analysis. Furthermore, we performed a comparison and protein-protein interaction analysis to identify causal proteins. We also analyzed the possible consequences of our discoveries by the given existing asthma drugs and their targets.
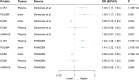
Results: Using Mendelian randomization analysis, we identified five protein-asthma pairs that were significant at the Bonferroni level (P < 6.35 × 10-5). Specifically, in plasma, we found that an increase of one standard deviation in IL1R1 and ECM1 was associated with an increased risk of asthma, while an increase in ADAM19 was found to be protective. The corresponding odds ratios were 1.03 (95% CI, 1.02-1.04), 1.00 (95% CI, 1.00-1.01), and 0.99 (95% CI, 0.98-0.99), respectively. In the brain, per 10-fold increase in ECM1 (OR, 1.05; 95% CI, 1.03-1.08) and PDLIM4 (OR, 1.05; 95% CI, 1.04-1.07) increased the risk of asthma. Bayesian co-localization found that ECM1 in the plasma (coloc.abf-PPH4 = 0.965) and in the brain (coloc.abf-PPH4 = 0.931) shared the same mutation with asthma. The target proteins of current asthma medications were found to interact with IL1R1. IL1R1 and PDLIM4 were validated in two replication cohorts.
Conclusion: Our integrative analysis revealed that asthma risk is causally affected by the levels of IL1R1, ECM1, and PDLIM4. The results suggest that these three proteins have the potential to be used as drug targets for asthma, and further investigation through clinical trials is needed.
Keywords: Mendelian randomization; asthma; drug target; neurogenic inflammation; protein quantitative trait loci.
Copyright © 2023 Wang, Wang, Yan, Liu and Xu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Potential drug targets for multiple sclerosis identified through Mendelian randomization analysis.Brain. 2023 Aug 1;146(8):3364-3372. doi: 10.1093/brain/awad070. Brain. 2023. PMID: 36864689 Free PMC article.
-
Identification of potential drug targets for allergic diseases from a genetic perspective: A mendelian randomization study.Clin Transl Allergy. 2024 Apr;14(4):e12350. doi: 10.1002/clt2.12350. Clin Transl Allergy. 2024. PMID: 38573314 Free PMC article.
-
Exploration of potential novel drug targets for diabetic retinopathy by plasma proteome screening.Sci Rep. 2024 May 22;14(1):11726. doi: 10.1038/s41598-024-62069-0. Sci Rep. 2024. PMID: 38778174 Free PMC article.
-
Potential therapeutic targets for membranous nephropathy: proteome-wide Mendelian randomization and colocalization analysis.Front Immunol. 2024 Apr 19;15:1342912. doi: 10.3389/fimmu.2024.1342912. eCollection 2024. Front Immunol. 2024. PMID: 38707900 Free PMC article.
-
Investigating potential drug targets for IgA nephropathy and membranous nephropathy through multi-queue plasma protein analysis: a Mendelian randomization study based on SMR and co-localization analysis.BioData Min. 2024 Nov 8;17(1):49. doi: 10.1186/s13040-024-00405-w. BioData Min. 2024. PMID: 39516845 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

