Cell type-specific delivery by modular envelope design
- PMID: 37612276
- PMCID: PMC10447438
- DOI: 10.1038/s41467-023-40788-8
Cell type-specific delivery by modular envelope design
Abstract
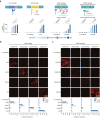
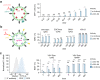
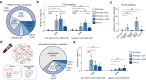
The delivery of genetic cargo remains one of the largest obstacles to the successful translation of experimental therapies, in large part due to the absence of targetable delivery vectors. Enveloped delivery modalities use viral envelope proteins, which determine tropism and induce membrane fusion. Here we develop DIRECTED (Delivery to Intended REcipient Cells Through Envelope Design), a modular platform that consists of separate fusion and targeting components. To achieve high modularity and programmable cell type specificity, we develop multiple strategies to recruit or immobilize antibodies on the viral envelope, including a chimeric antibody binding protein and a SNAP-tag enabling the use of antibodies or other proteins as targeting molecules. Moreover, we show that fusogens from multiple viral families are compatible with DIRECTED and that DIRECTED components can target multiple delivery chassis (e.g., lentivirus and MMLV gag) to specific cell types, including primary human T cells in PBMCs and whole blood.
© 2023. Springer Nature Limited.
Conflict of interest statement
D.S. and F.Z. are coinventors on a pending patent application (PCT/US22/52871) related to this work filed by the Broad Institute and MIT. M.J.F. reports receiving speaker’s bureau honoraria from Pfizer. F.Z. is a scientific advisor and cofounder of Editas Medicine, Beam Therapeutics, Pairwise Plants, Arbor Biotechnologies, and Aera Therapeutics. F.Z. is a scientific advisor for Octant. The remaining authors declare no competing interests.
Figures



Similar articles
-
Efficient and Highly Specific Gene Transfer Using Mutated Lentiviral Vectors Redirected with Bispecific Antibodies.mBio. 2020 Jan 21;11(1):e02990-19. doi: 10.1128/mBio.02990-19. mBio. 2020. PMID: 31964730 Free PMC article.
-
Cell-specific targeting of lentiviral vectors mediated by fusion proteins derived from Sindbis virus, vesicular stomatitis virus, or avian sarcoma/leukosis virus.Retrovirology. 2010 Jan 25;7:3. doi: 10.1186/1742-4690-7-3. Retrovirology. 2010. PMID: 20100344 Free PMC article.
-
Conformation-specific antibodies targeting the trimer-of-hairpins motif of the human T-cell leukemia virus type 1 transmembrane glycoprotein recognize the viral envelope but fail to neutralize viral entry.J Virol. 2007 Jun;81(11):6019-31. doi: 10.1128/JVI.02544-06. Epub 2007 Mar 21. J Virol. 2007. PMID: 17376912 Free PMC article.
-
Altering the tropism of lentiviral vectors through pseudotyping.Curr Gene Ther. 2005 Aug;5(4):387-98. doi: 10.2174/1566523054546224. Curr Gene Ther. 2005. PMID: 16101513 Free PMC article. Review.
-
Receptors and tropisms of envelope viruses.Curr Opin Virol. 2011 Jul;1(1):13-8. doi: 10.1016/j.coviro.2011.05.001. Curr Opin Virol. 2011. PMID: 21804908 Free PMC article. Review.
Cited by
-
Molecular Engineering of Virus Tropism.Int J Mol Sci. 2024 Oct 15;25(20):11094. doi: 10.3390/ijms252011094. Int J Mol Sci. 2024. PMID: 39456875 Free PMC article. Review.
-
A nanobody interaction with SARS-COV-2 Spike allows the versatile targeting of lentivirus vectors.J Virol. 2024 Sep 17;98(9):e0079524. doi: 10.1128/jvi.00795-24. Epub 2024 Aug 29. J Virol. 2024. PMID: 39207135 Free PMC article.
-
Emerging trends in virus and virus-like particle gene therapy delivery to the brain.Mol Ther Nucleic Acids. 2024 Jul 19;35(3):102280. doi: 10.1016/j.omtn.2024.102280. eCollection 2024 Sep 10. Mol Ther Nucleic Acids. 2024. PMID: 39206077 Free PMC article. Review.
-
Advances in application of CRISPR-Cas13a system.Front Cell Infect Microbiol. 2024 Feb 19;14:1291557. doi: 10.3389/fcimb.2024.1291557. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 38524179 Free PMC article. Review.
-
Bottom-up synthetic immunology.Nat Nanotechnol. 2024 Nov;19(11):1587-1596. doi: 10.1038/s41565-024-01744-9. Epub 2024 Aug 26. Nat Nanotechnol. 2024. PMID: 39187581 Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

