Global Distribution of Founder Variants Associated with Non-Syndromic Hearing Impairment
- PMID: 36833326
- PMCID: PMC9957346
- DOI: 10.3390/genes14020399
Global Distribution of Founder Variants Associated with Non-Syndromic Hearing Impairment
Abstract
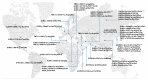
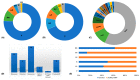
The genetic etiology of non-syndromic hearing impairment (NSHI) is highly heterogeneous with over 124 distinct genes identified. The wide spectrum of implicated genes has challenged the implementation of molecular diagnosis with equal clinical validity in all settings. Differential frequencies of allelic variants in the most common NSHI causal gene, gap junction beta 2 (GJB2), has been described as stemming from the segregation of a founder variant and/or spontaneous germline variant hot spots. We aimed to systematically review the global distribution and provenance of founder variants associated with NSHI. The study protocol was registered on PROSPERO, the International Prospective Register of Systematic Reviews, with the registration number "CRD42020198573". Data from 52 reports, involving 27,959 study participants from 24 countries, reporting 56 founder pathogenic or likely pathogenic (P/LP) variants in 14 genes (GJB2, GJB6, GSDME, TMC1, TMIE, TMPRSS3, KCNQ4, PJVK, OTOF, EYA4, MYO15A, PDZD7, CLDN14, and CDH23), were reviewed. Varied number short tandem repeats (STRs) and single nucleotide polymorphisms (SNPs) were used for haplotype analysis to identify the shared ancestral informative markers in a linkage disequilibrium and variants' origins, age estimates, and common ancestry computations in the reviewed reports. Asia recorded the highest number of NSHI founder variants (85.7%; 48/56), with variants in all 14 genes, followed by Europe (16.1%; 9/56). GJB2 had the highest number of ethnic-specific P/LP founder variants. This review reports on the global distribution of NSHI founder variants and relates their evolution to population migration history, bottleneck events, and demographic changes in populations linked with the early evolution of deleterious founder alleles. International migration and regional and cultural intermarriage, coupled to rapid population growth, may have contributed to re-shaping the genetic architecture and structural dynamics of populations segregating these pathogenic founder variants. We have highlighted and showed the paucity of data on hearing impairment (HI) variants in Africa, establishing unexplored opportunities in genetic traits.
Keywords: founder variant; gab junction beta 2 (GJB2); genetics; global populations; hearing impairment; non-syndromic.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Connexin Genes Variants Associated with Non-Syndromic Hearing Impairment: A Systematic Review of the Global Burden.Life (Basel). 2020 Oct 28;10(11):258. doi: 10.3390/life10110258. Life (Basel). 2020. PMID: 33126609 Free PMC article. Review.
-
Epidemiology, etiology, genetic variants in non- syndromic hearing loss in Iran: A systematic review and meta-analysis.Int J Pediatr Otorhinolaryngol. 2023 May;168:111512. doi: 10.1016/j.ijporl.2023.111512. Epub 2023 Mar 29. Int J Pediatr Otorhinolaryngol. 2023. PMID: 37086676 Review.
-
Genetic etiology of non-syndromic hearing loss in Europe.Hum Genet. 2022 Apr;141(3-4):683-696. doi: 10.1007/s00439-021-02425-6. Epub 2022 Jan 19. Hum Genet. 2022. PMID: 35044523 Review.
-
GJB2 and GJB6 mutations in 165 Danish patients showing non-syndromic hearing impairment.Genet Test. 2004 Summer;8(2):181-4. doi: 10.1089/gte.2004.8.181. Genet Test. 2004. PMID: 15345117
-
Spectrum and frequencies of non GJB2 gene mutations in Czech patients with early non-syndromic hearing loss detected by gene panel NGS and whole-exome sequencing.Clin Genet. 2020 Dec;98(6):548-554. doi: 10.1111/cge.13839. Epub 2020 Sep 27. Clin Genet. 2020. PMID: 32860223
Cited by
-
The GJB2 (Cx26) Gene Variants in Patients with Hearing Impairment in the Baikal Lake Region (Russia).Genes (Basel). 2023 Apr 28;14(5):1001. doi: 10.3390/genes14051001. Genes (Basel). 2023. PMID: 37239361 Free PMC article.
-
Novel, pathogenic insertion variant of GSDME associates with autosomal dominant hearing loss in a large Chinese pedigree.J Cell Mol Med. 2024 Jan;28(1):e18004. doi: 10.1111/jcmm.18004. Epub 2023 Oct 20. J Cell Mol Med. 2024. PMID: 37864300 Free PMC article.
-
An overload of missense variants in the OTOG gene may drive a higher prevalence of familial Meniere disease in the European population.Hum Genet. 2024 Mar;143(3):423-435. doi: 10.1007/s00439-024-02643-8. Epub 2024 Mar 22. Hum Genet. 2024. PMID: 38519595 Free PMC article.
-
The historical demography of the Martha's Vineyard signing community.J Deaf Stud Deaf Educ. 2024 Jun 24;29(3):295-321. doi: 10.1093/deafed/enad058. J Deaf Stud Deaf Educ. 2024. PMID: 38287681 Free PMC article.
-
Comprehensive analysis, diagnosis, prognosis, and cordycepin (CD) regulations for GSDME expressions in pan-cancers.Cancer Cell Int. 2024 Aug 8;24(1):279. doi: 10.1186/s12935-024-03467-2. Cancer Cell Int. 2024. PMID: 39118110 Free PMC article.
References
-
- Bliznetz E.A., Lalayants M.R., Markova T.G., Balanovsky O.P., Balanovska E.V., Skhalyakho R.A., Pocheshkhova E.A., Nikitina N.V., Voronin S.V., Kudryashova E.K. Update of the GJB2/DFNB1 Mutation Spectrum in Russia: A Founder Ingush Mutation Del (GJB2-D13S175) Is the Most Frequent among Other Large Deletions. J. Hum. Genet. 2017;62:789–795. doi: 10.1038/jhg.2017.42. - DOI - PMC - PubMed
-
- Mikstiene V., Jakaitiene A., Byckova J., Gradauskiene E., Preiksaitiene E., Burnyte B., Tumiene B., Matuleviciene A., Ambrozaityte L., Uktveryte I. The High Frequency of GJB2 Gene Mutation c. 313_326del14 Suggests Its Possible Origin in Ancestors of Lithuanian Population. BMC Genet. 2016;17:1–12. doi: 10.1186/s12863-016-0354-9. - DOI - PMC - PubMed
-
- Borck G., Rainshtein L., Hellman-Aharony S., Volk A.E., Friedrich K., Taub E., Magal N., Kanaan M., Kubisch C., Shohat M. High Frequency of Autosomal-Recessive DFNB59 Hearing Loss in an Isolated Arab Population in Israel. Clin. Genet. 2012;82:271–276. doi: 10.1111/j.1399-0004.2011.01741.x. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

