MetaProD: A Highly-Configurable Mass Spectrometry Analyzer for Multiplexed Proteomic and Metaproteomic Data
- PMID: 36688801
- PMCID: PMC9903327
- DOI: 10.1021/acs.jproteome.2c00614
MetaProD: A Highly-Configurable Mass Spectrometry Analyzer for Multiplexed Proteomic and Metaproteomic Data
Abstract
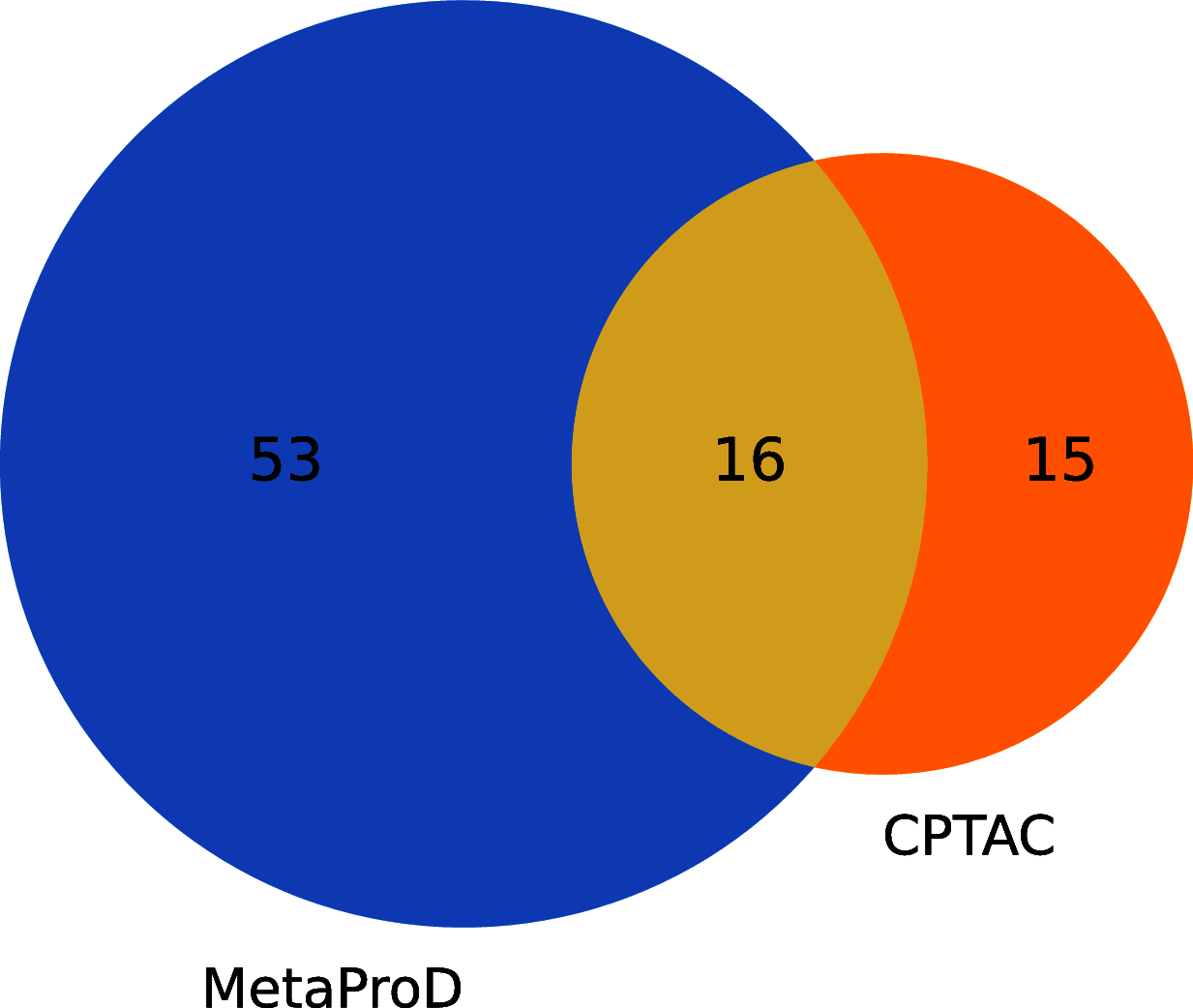
The microbiome has been shown to be important for human health because of its influence on disease and the immune response. Mass spectrometry is an important tool for evaluating protein expression and species composition in the microbiome but is technically challenging and time-consuming. Multiplexing has emerged as a way to make spectrometry workflows faster while improving results. Here, we present MetaProD (MetaProteomics in Django) as a highly configurable metaproteomic data analysis pipeline supporting label-free and multiplexed mass spectrometry. The pipeline is open-source, uses fully open-source tools, and is integrated with Django to offer a web-based interface for configuration and data access. Benchmarking of MetaProD using multiple metaproteomics data sets showed that MetaProD achieved fast and efficient identification of peptides and proteins. Application of MetaProD to a multiplexed cancer data set resulted in identification of more differentially expressed human proteins in cancer tissues versus healthy tissues as compared to previous studies; in addition, MetaProD identified bacterial proteins in those samples, some of which are differentially abundant.
Keywords: bacterial proteins; differentially expressed proteins; isobaric labeling; mass spectrometry; metaproteomics; multiplexing; proteomics.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

Similar articles
-
Using high-abundance proteins as guides for fast and effective peptide/protein identification from human gut metaproteomic data.Microbiome. 2021 Apr 1;9(1):80. doi: 10.1186/s40168-021-01035-8. Microbiome. 2021. PMID: 33795009 Free PMC article.
-
MetaProClust-MS1: an MS1 Profiling Approach for Large-Scale Microbiome Screening.mSystems. 2022 Aug 30;7(4):e0038122. doi: 10.1128/msystems.00381-22. Epub 2022 Aug 11. mSystems. 2022. PMID: 35950762 Free PMC article.
-
Optimizing metaproteomics database construction: lessons from a study of the vaginal microbiome.mSystems. 2023 Aug 31;8(4):e0067822. doi: 10.1128/msystems.00678-22. Epub 2023 Jun 23. mSystems. 2023. PMID: 37350639 Free PMC article.
-
Metaproteomics as a Complementary Approach to Gut Microbiota in Health and Disease.Front Chem. 2017 Jan 26;5:4. doi: 10.3389/fchem.2017.00004. eCollection 2017. Front Chem. 2017. PMID: 28184370 Free PMC article. Review.
-
Metaproteomic and Metabolomic Approaches for Characterizing the Gut Microbiome.Proteomics. 2019 Aug;19(16):e1800363. doi: 10.1002/pmic.201800363. Epub 2019 Jul 31. Proteomics. 2019. PMID: 31321880 Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

