Emerging Roles of NDUFS8 Located in Mitochondrial Complex I in Different Diseases
- PMID: 36557887
- PMCID: PMC9783039
- DOI: 10.3390/molecules27248754
Emerging Roles of NDUFS8 Located in Mitochondrial Complex I in Different Diseases
Abstract
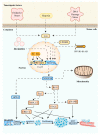
NADH:ubiquinone oxidoreductase core subunit S8 (NDUFS8) is an essential core subunit and component of the iron-sulfur (FeS) fragment of mitochondrial complex I directly involved in the electron transfer process and energy metabolism. Pathogenic variants of the NDUFS8 are relevant to infantile-onset and severe diseases, including Leigh syndrome, cancer, and diabetes mellitus. With over 1000 nuclear genes potentially causing a mitochondrial disorder, the current diagnostic approach requires targeted molecular analysis, guided by a combination of clinical and biochemical features. Currently, there are only several studies on pathogenic variants of the NDUFS8 in Leigh syndrome, and a lack of literature on its precise mechanism in cancer and diabetes mellitus exists. Therefore, NDUFS8-related diseases should be extensively explored and precisely diagnosed at the molecular level with the application of next-generation sequencing technologies. A more distinct comprehension will be needed to shed light on NDUFS8 and its related diseases for further research. In this review, a comprehensive summary of the current knowledge about NDUFS8 structural function, its pathogenic mutations in Leigh syndrome, as well as its underlying roles in cancer and diabetes mellitus is provided, offering potential pathogenesis, progress, and therapeutic target of different diseases. We also put forward some problems and solutions for the following investigations.
Keywords: Leigh syndrome; NDUFS8; cancer; diabetes mellitus; metabolism; mitochondrial complex I.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Late-onset Leigh syndrome in a patient with mitochondrial complex I NDUFS8 mutations.Neurology. 2004 May 25;62(10):1899-901. doi: 10.1212/01.wnl.0000125251.56131.65. Neurology. 2004. PMID: 15159508 Free PMC article. Review.
-
Mutant NDUFS3 subunit of mitochondrial complex I causes Leigh syndrome.J Med Genet. 2004 Jan;41(1):14-7. doi: 10.1136/jmg.2003.014316. J Med Genet. 2004. PMID: 14729820 Free PMC article.
-
TAT-Conjugated NDUFS8 Can Be Transduced into Mitochondria in a Membrane-Potential-Independent Manner and Rescue Complex I Deficiency.Int J Mol Sci. 2021 Jun 17;22(12):6524. doi: 10.3390/ijms22126524. Int J Mol Sci. 2021. PMID: 34204592 Free PMC article.
-
Leigh syndrome associated with mitochondrial complex I deficiency due to novel mutations In NDUFV1 and NDUFS2.Gene. 2013 Mar 1;516(1):162-7. doi: 10.1016/j.gene.2012.12.024. Epub 2012 Dec 22. Gene. 2013. PMID: 23266820
-
NDUFS6 related Leigh syndrome: a case report and review of the literature.J Hum Genet. 2019 Jul;64(7):637-645. doi: 10.1038/s10038-019-0594-4. Epub 2019 Apr 4. J Hum Genet. 2019. PMID: 30948790 Review.
Cited by
-
Revealing the potential role of hub metabolism-related genes and their correlation with immune cells in acute ischemic stroke.IET Syst Biol. 2024 Aug;18(4):129-142. doi: 10.1049/syb2.12095. Epub 2024 Jun 8. IET Syst Biol. 2024. PMID: 38850201 Free PMC article.
-
Western diet-induced obesity results in brain mitochondrial dysfunction in female Ossabaw swine.Front Mol Neurosci. 2023 Dec 14;16:1320879. doi: 10.3389/fnmol.2023.1320879. eCollection 2023. Front Mol Neurosci. 2023. PMID: 38163062 Free PMC article.
-
Dose- and Time-Dependent Effects of Radiofrequency Electromagnetic Field on Adipose Tissue: Implications of Thermoregulation and Mitochondrial Signaling.Int J Mol Sci. 2023 Jun 25;24(13):10628. doi: 10.3390/ijms241310628. Int J Mol Sci. 2023. PMID: 37445806 Free PMC article.
-
The requirement of the mitochondrial protein NDUFS8 for angiogenesis.Cell Death Dis. 2024 Apr 9;15(4):253. doi: 10.1038/s41419-024-06636-3. Cell Death Dis. 2024. PMID: 38594244 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

