Genetic regulatory and biological implications of the 10q24.32 schizophrenia risk locus
- PMID: 36152315
- PMCID: PMC10115178
- DOI: 10.1093/brain/awac352
Genetic regulatory and biological implications of the 10q24.32 schizophrenia risk locus
Abstract
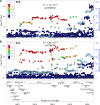
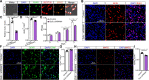
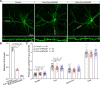
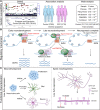
Genome-wide association studies have identified 10q24.32 as a robust schizophrenia risk locus. Here we identify a regulatory variant (rs10786700) that disrupts binding of transcription factors at 10q24.32. We independently confirmed the association between rs10786700 and schizophrenia in a large Chinese cohort (n = 11 547) and uncovered the biological mechanism underlying this association. We found that rs10786700 resides in a super-enhancer element that exhibits dynamic activity change during the development process and that the risk allele (C) of rs10786700 conferred significant lower enhancer activity through enhancing binding affinity to repressor element-1 silencing transcription factor (REST). CRISPR-Cas9-mediated genome editing identified SUFU as a potential target gene by which rs10786700 might exert its risk effect on schizophrenia, as deletion of rs10786700 downregulated SUFU expression. We further investigated the role of Sufu in neurodevelopment and found that Sufu knockdown inhibited proliferation of neural stem cells and neurogenesis, affected molecular pathways (including neurodevelopment-related pathways, PI3K-Akt and ECM-receptor interaction signalling pathways) associated with schizophrenia and altered the density of dendritic spines. These results reveal that the functional risk single nucleotide polymorphism rs10786700 at 10q24.32 interacts with REST synergistically to regulate expression of SUFU, a novel schizophrenia risk gene which is involved in schizophrenia pathogenesis by affecting neurodevelopment and spine morphogenesis.
Keywords: SUFU; dendritic spine density; functional risk variant; rs10786700; schizophrenia.
© The Author(s) 2022. Published by Oxford University Press on behalf of the Guarantors of Brain.
Conflict of interest statement
The authors report no biomedical financial interests or potential conflicts of interest.
Figures



Similar articles
-
Regulatory Variant rs2535629 in ITIH3 Intron Confers Schizophrenia Risk By Regulating CTCF Binding and SFMBT1 Expression.Adv Sci (Weinh). 2022 Feb;9(6):e2104786. doi: 10.1002/advs.202104786. Epub 2022 Jan 2. Adv Sci (Weinh). 2022. PMID: 34978167 Free PMC article.
-
Functional variant rs2270363 on 16p13.3 confers schizophrenia risk by regulating NMRAL1.Brain. 2022 Jul 29;145(7):2569-2585. doi: 10.1093/brain/awac020. Brain. 2022. PMID: 35094059 Free PMC article.
-
Regulatory variants at 2q33.1 confer schizophrenia risk by modulating distal gene TYW5 expression.Brain. 2022 Apr 18;145(2):770-786. doi: 10.1093/brain/awab357. Brain. 2022. PMID: 34581804 Free PMC article.
-
The schizophrenia risk gene ZNF804A: clinical associations, biological mechanisms and neuronal functions.Mol Psychiatry. 2017 Jul;22(7):944-953. doi: 10.1038/mp.2017.19. Epub 2017 Mar 14. Mol Psychiatry. 2017. PMID: 28289284 Review.
-
A re-review of the association between the NOTCH4 locus and schizophrenia.Am J Med Genet B Neuropsychiatr Genet. 2012 Jul;159B(5):477-83. doi: 10.1002/ajmg.b.32050. Epub 2012 Apr 9. Am J Med Genet B Neuropsychiatr Genet. 2012. PMID: 22488909 Review.
Cited by
-
Endothelial Arid1a deletion disrupts the balance among angiogenesis, neurogenesis and gliogenesis in the developing brain.Cell Prolif. 2023 May;56(5):e13447. doi: 10.1111/cpr.13447. Epub 2023 Mar 13. Cell Prolif. 2023. PMID: 36916004 Free PMC article.
-
Integrating Genetic and Transcriptomic Data to Identify Genes Underlying Obesity Risk Loci.medRxiv [Preprint]. 2024 Jun 12:2024.06.11.24308730. doi: 10.1101/2024.06.11.24308730. medRxiv. 2024. PMID: 38903089 Free PMC article. Preprint.
-
Pathogenic role of super-enhancers as potential therapeutic targets in lung cancer.Front Pharmacol. 2024 Apr 12;15:1383580. doi: 10.3389/fphar.2024.1383580. eCollection 2024. Front Pharmacol. 2024. PMID: 38681203 Free PMC article. Review.
-
Massively parallel characterization of psychiatric disorder-associated and cell-type-specific regulatory elements in the developing human cortex.bioRxiv [Preprint]. 2023 Feb 16:2023.02.15.528663. doi: 10.1101/2023.02.15.528663. bioRxiv. 2023. Update in: Science. 2024 May 24;384(6698):eadh0559. doi: 10.1126/science.adh0559. PMID: 36824845 Free PMC article. Updated. Preprint.
References
-
- Birnbaum R, Weinberger DR. Genetic insights into the neurodevelopmental origins of schizophrenia. Nat Rev Neurosci. 2017;18:727–740. - PubMed
-
- Weinberger DR. Implications of normal brain development for the pathogenesis of schizophrenia. Arch Gen Psychiatry.1987;44:660–669. - PubMed
-
- Thaker GK, Carpenter WT. Advances in schizophrenia. Nat Med. 2001;7:667–671. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

