A Comprehensive Overview of Globally Approved JAK Inhibitors
- PMID: 35631587
- PMCID: PMC9146299
- DOI: 10.3390/pharmaceutics14051001
A Comprehensive Overview of Globally Approved JAK Inhibitors
Abstract
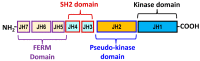
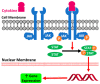
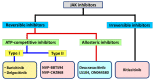
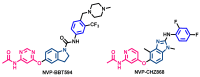
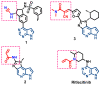
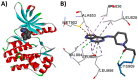
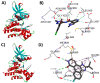
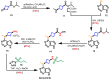
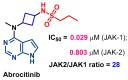
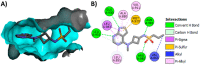
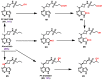
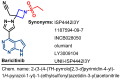
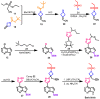
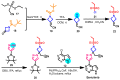
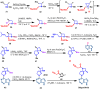
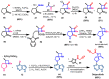
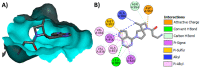
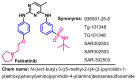
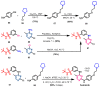
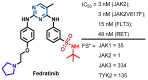
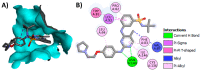
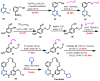
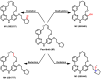
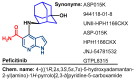
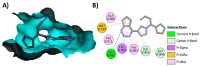
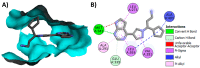
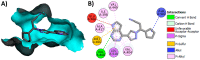
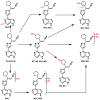
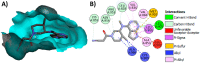
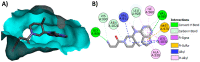
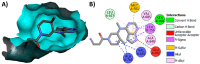
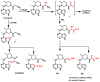
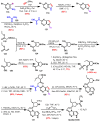
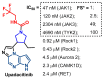
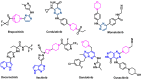
Janus kinase (JAK) is a family of cytoplasmic non-receptor tyrosine kinases that includes four members, namely JAK1, JAK2, JAK3, and TYK2. The JAKs transduce cytokine signaling through the JAK-STAT pathway, which regulates the transcription of several genes involved in inflammatory, immune, and cancer conditions. Targeting the JAK family kinases with small-molecule inhibitors has proved to be effective in the treatment of different types of diseases. In the current review, eleven of the JAK inhibitors that received approval for clinical use have been discussed. These drugs are abrocitinib, baricitinib, delgocitinib, fedratinib, filgotinib, oclacitinib, pacritinib, peficitinib, ruxolitinib, tofacitinib, and upadacitinib. The aim of the current review was to provide an integrated overview of the chemical and pharmacological data of the globally approved JAK inhibitors. The synthetic routes of the eleven drugs were described. In addition, their inhibitory activities against different kinases and their pharmacological uses have also been explained. Moreover, their crystal structures with different kinases were summarized, with a primary focus on their binding modes and interactions. The proposed metabolic pathways and metabolites of these drugs were also illustrated. To sum up, the data in the current review could help in the design of new JAK inhibitors with potential therapeutic benefits in inflammatory and autoimmune diseases.
Keywords: JAK; binding mode/interactions; kinase inhibitory activity; pharmacological uses; synthesis.
Conflict of interest statement
Authors declared that there is no conflict of interest and have approved the article.
Figures
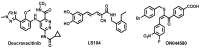
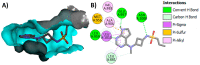



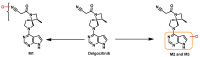






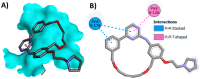







Similar articles
-
First Approval of Pacritinib as a Selective Janus Associated Kinase-2 Inhibitor for the Treatment of Patients with Myelofibrosis.Anticancer Agents Med Chem. 2023;23(12):1355-1360. doi: 10.2174/1871520623666230320120915. Anticancer Agents Med Chem. 2023. PMID: 36959157
-
Janus kinase (JAK) inhibitors in the treatment of neoplastic and inflammatory disorders.Pharmacol Res. 2022 Sep;183:106362. doi: 10.1016/j.phrs.2022.106362. Epub 2022 Jul 22. Pharmacol Res. 2022. PMID: 35878738 Review.
-
Janus kinase (JAK) inhibitors in the treatment of inflammatory and neoplastic diseases.Pharmacol Res. 2016 Sep;111:784-803. doi: 10.1016/j.phrs.2016.07.038. Epub 2016 Jul 26. Pharmacol Res. 2016. PMID: 27473820 Review.
-
JAK inhibitors for rheumatoid arthritis.Expert Opin Investig Drugs. 2023 Apr;32(4):333-344. doi: 10.1080/13543784.2023.2199919. Epub 2023 Apr 7. Expert Opin Investig Drugs. 2023. PMID: 37014106 Review.
-
Deucravacitinib is an allosteric TYK2 protein kinase inhibitor FDA-approved for the treatment of psoriasis.Pharmacol Res. 2023 Mar;189:106642. doi: 10.1016/j.phrs.2022.106642. Epub 2023 Feb 6. Pharmacol Res. 2023. PMID: 36754102 Review.
Cited by
-
Assessment of the Safety of Tofacitinib Among Patients With Psoriasis: A Systematic Review and Meta-Analysis.Cureus. 2024 Sep 25;16(9):e70196. doi: 10.7759/cureus.70196. eCollection 2024 Sep. Cureus. 2024. PMID: 39463512 Free PMC article. Review.
-
JAK inhibition decreases the autoimmune burden in Down syndrome.medRxiv [Preprint]. 2024 Oct 16:2024.06.13.24308783. doi: 10.1101/2024.06.13.24308783. medRxiv. 2024. Update in: Elife. 2024 Dec 31;13:RP99323. doi: 10.7554/eLife.99323. PMID: 38946973 Free PMC article. Updated. Preprint.
-
Discovery of GLPG3667, a Selective ATP Competitive Tyrosine Kinase 2 Inhibitor for the Treatment of Autoimmune Diseases.J Med Chem. 2024 Jun 13;67(11):8545-8568. doi: 10.1021/acs.jmedchem.4c00769. Epub 2024 May 28. J Med Chem. 2024. PMID: 38805213 Free PMC article. Clinical Trial.
-
Inflammation as a cause of acute myocardial infarction in patients with myeloproliferative neoplasm.World J Cardiol. 2024 Feb 26;16(2):58-63. doi: 10.4330/wjc.v16.i2.58. World J Cardiol. 2024. PMID: 38456066 Free PMC article.
-
"Oh, Dear We Are in Tribble": An Overview of the Oncogenic Functions of Tribbles 1.Cancers (Basel). 2024 May 16;16(10):1889. doi: 10.3390/cancers16101889. Cancers (Basel). 2024. PMID: 38791967 Free PMC article. Review.
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

