Targeting autophagy using small-molecule compounds to improve potential therapy of Parkinson's disease
- PMID: 34729301
- PMCID: PMC8546670
- DOI: 10.1016/j.apsb.2021.02.016
Targeting autophagy using small-molecule compounds to improve potential therapy of Parkinson's disease
Abstract
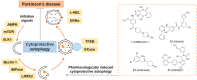
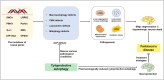
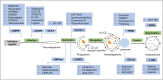
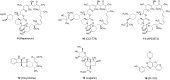
Parkinson's disease (PD), known as one of the most universal neurodegenerative diseases, is a serious threat to the health of the elderly. The current treatment has been demonstrated to relieve symptoms, and the discovery of new small-molecule compounds has been regarded as a promising strategy. Of note, the homeostasis of the autolysosome pathway (ALP) is closely associated with PD, and impaired autophagy may cause the death of neurons and thereby accelerating the progress of PD. Thus, pharmacological targeting autophagy with small-molecule compounds has been drawn a rising attention so far. In this review, we focus on summarizing several autophagy-associated targets, such as AMPK, mTORC1, ULK1, IMPase, LRRK2, beclin-1, TFEB, GCase, ERRα, C-Abelson, and as well as their relevant small-molecule compounds in PD models, which will shed light on a clue on exploiting more potential targeted small-molecule drugs tracking PD treatment in the near future.
Keywords: 3-MA, 3-methyladenine; 5-HT2A, Serotonin 2A; 5-HT2C, serotonin 2C; A2A, adenosine 2A; AADC, aromatic amino acid decarboxylase; ALP, autophagy-lysosomal pathway; AMPK, 5ʹAMP-activated protein kinase; ATG, autophagy related protein; ATP13A2, ATPase cation transporting 13A2; ATTEC, autophagosome-tethering compound; AUC, the area under the curve; AUTAC, autophagy targeting chimera; Autophagy; BAF, bafilomycinA1; BBB, blood−brain barrier; CL, clearance rate; CMA, chaperone-mediated autophagy; CNS, central nervous system; COMT, catechol-O-methyltransferase; DA, dopamine; DAT, dopamine transporter; DJ-1, Parkinson protein 7; DR, dopamine receptor; ER, endoplasmic reticulum; ERRα, estrogen-related receptor alpha; F, oral bioavailability; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; GBA, glucocerebrosidase β acid; GWAS, genome-wide association study; HDAC6, histone deacetylase 6; HSC70, heat shock cognate 71 kDa protein; HSPA8, heat shock 70 kDa protein 8; IMPase, inositol monophosphatase; IPPase, inositol polyphosphate 1-phosphatase; KI, knockin; LAMP2A, lysosome-associated membrane protein 2 A; LC3, light chain 3; LIMP-2, lysosomal integrated membrane protein-2; LRRK2, leucine-rich repeat sequence kinase 2; LRS, leucyl-tRNA synthetase; LUHMES, lund human mesencephalic; Lamp2a, type 2A lysosomal-associated membrane protein; MAO-B, monoamine oxidase B; MPP+, 1-methyl-4-phenylpyridinium; MPTP, 1-methyl-4-phenyl-1,2,5,6-tetrahydropyridine; MYCBP2, MYC-binding protein 2; NMDA, N-methyl-d-aspartic acid; ONRs, orphan nuclear receptors; PD therapy; PD, Parkinson's disease; PDE4, phosphodiesterase 4; PI3K, phosphatidylinositol 3-kinase; PI3P, phosphatidylinositol 3-phosphate; PINK1, PTEN-induced kinase 1; PLC, phospholipase C; PREP, prolyl oligopeptidase; Parkin, parkin RBR E3 ubiquitin−protein ligase; Parkinson's disease (PD); ROS, reactive oxygen species; SAR, structure–activity relationship; SAS, solvent accessible surface; SN, substantia nigra; SNCA, α-synuclein gene; SYT11, synaptotagmin 11; Small-molecule compound; TFEB, transcription factor EB; TSC2, tuberous sclerosis complex 2; Target; ULK1, UNC-51-like kinase 1; UPS, ubiquitin−proteasome system; mAChR, muscarinic acetylcholine receptor; mTOR, the mammalian target of rapamycin; α-syn, α-synuclein.
© 2021 Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences. Production and hosting by Elsevier B.V.
Conflict of interest statement
The authors have no conflicts of interest to declare.
Figures



Similar articles
-
Age-dependent accumulation of oligomeric SNCA/α-synuclein from impaired degradation in mutant LRRK2 knockin mouse model of Parkinson disease: role for therapeutic activation of chaperone-mediated autophagy (CMA).Autophagy. 2020 Feb;16(2):347-370. doi: 10.1080/15548627.2019.1603545. Epub 2019 Apr 14. Autophagy. 2020. PMID: 30983487 Free PMC article.
-
Impaired autophagy and APP processing in Alzheimer's disease: The potential role of Beclin 1 interactome.Prog Neurobiol. 2013 Jul-Aug;106-107:33-54. doi: 10.1016/j.pneurobio.2013.06.002. Epub 2013 Jul 1. Prog Neurobiol. 2013. PMID: 23827971 Review.
-
The ménage à trois of autophagy, lipid droplets and liver disease.Autophagy. 2022 Jan;18(1):50-72. doi: 10.1080/15548627.2021.1895658. Epub 2021 Apr 2. Autophagy. 2022. PMID: 33794741 Free PMC article. Review.
-
Organelle-specific autophagy in inflammatory diseases: a potential therapeutic target underlying the quality control of multiple organelles.Autophagy. 2021 Feb;17(2):385-401. doi: 10.1080/15548627.2020.1725377. Epub 2020 Feb 12. Autophagy. 2021. PMID: 32048886 Free PMC article. Review.
-
Copper metabolism in cell death and autophagy.Autophagy. 2023 Aug;19(8):2175-2195. doi: 10.1080/15548627.2023.2200554. Epub 2023 Apr 16. Autophagy. 2023. PMID: 37055935 Free PMC article. Review.
Cited by
-
Alpha-Pinene Decreases the Elevated Levels of Astrogliosis, Pyroptosis, and Autophagy Markers in the Hippocampus Triggered by Kainate in a Rat Model of Temporal Lobe Epilepsy.Mol Neurobiol. 2025 Feb;62(2):2264-2276. doi: 10.1007/s12035-024-04407-x. Epub 2024 Aug 3. Mol Neurobiol. 2025. PMID: 39096444
-
Autophagy in Parkinson's Disease.Biomolecules. 2023 Sep 22;13(10):1435. doi: 10.3390/biom13101435. Biomolecules. 2023. PMID: 37892117 Free PMC article. Review.
-
Loss of SARM1 ameliorates secondary thalamic neurodegeneration after cerebral infarction.J Cereb Blood Flow Metab. 2024 Feb;44(2):224-238. doi: 10.1177/0271678X231210694. Epub 2023 Oct 28. J Cereb Blood Flow Metab. 2024. PMID: 37898107 Free PMC article.
-
The Rare Marine Bioactive Compounds in Neurological Disorders and Diseases: Is the Blood-Brain Barrier an Obstacle or a Target?Mar Drugs. 2023 Jul 18;21(7):406. doi: 10.3390/md21070406. Mar Drugs. 2023. PMID: 37504937 Free PMC article. Review.
-
C9orf72 gene networks in the human brain correlate with cortical thickness in C9-FTD and implicate vulnerable cell types.Front Neurosci. 2024 Feb 26;18:1258996. doi: 10.3389/fnins.2024.1258996. eCollection 2024. Front Neurosci. 2024. PMID: 38469573 Free PMC article.
References
-
- Kalia L.V., Lang A.E. Parkinson's disease. Lancet. 2015;386:896–912. - PubMed
-
- Beaulieu J.M., Gainetdinov R.R. The physiology, signaling, and pharmacology of dopamine receptors. Pharmacol Rev. 2011;63:182–217. - PubMed
-
- Doty R.L. Olfactory dysfunction in Parkinson disease. Nat Rev Neurol. 2012;8:329–339. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

