Screening Potential Drugs for COVID-19 Based on Bound Nuclear Norm Regularization
- PMID: 34691157
- PMCID: PMC8529063
- DOI: 10.3389/fgene.2021.749256
Screening Potential Drugs for COVID-19 Based on Bound Nuclear Norm Regularization
Abstract
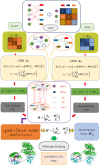
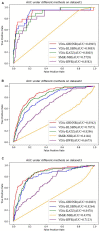
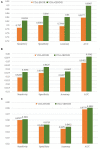
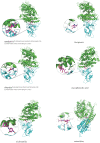
The novel coronavirus pneumonia COVID-19 infected by SARS-CoV-2 has attracted worldwide attention. It is urgent to find effective therapeutic strategies for stopping COVID-19. In this study, a Bounded Nuclear Norm Regularization (BNNR) method is developed to predict anti-SARS-CoV-2 drug candidates. First, three virus-drug association datasets are compiled. Second, a heterogeneous virus-drug network is constructed. Third, complete genomic sequences and Gaussian association profiles are integrated to compute virus similarities; chemical structures and Gaussian association profiles are integrated to calculate drug similarities. Fourth, a BNNR model based on kernel similarity (VDA-GBNNR) is proposed to predict possible anti-SARS-CoV-2 drugs. VDA-GBNNR is compared with four existing advanced methods under fivefold cross-validation. The results show that VDA-GBNNR computes better AUCs of 0.8965, 0.8562, and 0.8803 on the three datasets, respectively. There are 6 anti-SARS-CoV-2 drugs overlapping in any two datasets, that is, remdesivir, favipiravir, ribavirin, mycophenolic acid, niclosamide, and mizoribine. Molecular dockings are conducted for the 6 small molecules and the junction of SARS-CoV-2 spike protein and human angiotensin-converting enzyme 2. In particular, niclosamide and mizoribine show higher binding energy of -8.06 and -7.06 kcal/mol with the junction, respectively. G496 and K353 may be potential key residues between anti-SARS-CoV-2 drugs and the interface junction. We hope that the predicted results can contribute to the treatment of COVID-19.
Keywords: FDA-approved drugs; SARS-CoV-2; bounded nuclear norm regularization; molecular docking; virus-drug association.
Copyright © 2021 Wang, Wang, Shen, Zhou and Peng.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The handling editor declared a past co-authorship with one of the author LZ.
Figures
Similar articles
-
Drug repositioning for SARS-CoV-2 by Gaussian kernel similarity bilinear matrix factorization.Front Microbiol. 2022 Dec 5;13:1062281. doi: 10.3389/fmicb.2022.1062281. eCollection 2022. Front Microbiol. 2022. PMID: 36545200 Free PMC article.
-
Discovery of Potential Therapeutic Drugs for COVID-19 Through Logistic Matrix Factorization With Kernel Diffusion.Front Microbiol. 2022 Feb 28;13:740382. doi: 10.3389/fmicb.2022.740382. eCollection 2022. Front Microbiol. 2022. PMID: 35295301 Free PMC article.
-
Prioritizing antiviral drugs against SARS-CoV-2 by integrating viral complete genome sequences and drug chemical structures.Sci Rep. 2021 Mar 18;11(1):6248. doi: 10.1038/s41598-021-83737-5. Sci Rep. 2021. PMID: 33737523 Free PMC article.
-
Identifying Effective Antiviral Drugs Against SARS-CoV-2 by Drug Repositioning Through Virus-Drug Association Prediction.Front Genet. 2020 Sep 16;11:577387. doi: 10.3389/fgene.2020.577387. eCollection 2020. Front Genet. 2020. PMID: 33193695 Free PMC article.
-
VDA-RWLRLS: An anti-SARS-CoV-2 drug prioritizing framework combining an unbalanced bi-random walk and Laplacian regularized least squares.Comput Biol Med. 2022 Jan;140:105119. doi: 10.1016/j.compbiomed.2021.105119. Epub 2021 Dec 7. Comput Biol Med. 2022. PMID: 34902608 Free PMC article.
Cited by
-
Identifying shared genetic loci between coronavirus disease 2019 and cardiovascular diseases based on cross-trait meta-analysis.Front Microbiol. 2022 Sep 15;13:993933. doi: 10.3389/fmicb.2022.993933. eCollection 2022. Front Microbiol. 2022. PMID: 36187959 Free PMC article.
-
Drug repositioning for SARS-CoV-2 by Gaussian kernel similarity bilinear matrix factorization.Front Microbiol. 2022 Dec 5;13:1062281. doi: 10.3389/fmicb.2022.1062281. eCollection 2022. Front Microbiol. 2022. PMID: 36545200 Free PMC article.
-
Neighborhood-based inference and restricted Boltzmann machine for microbe and drug associations prediction.PeerJ. 2022 Aug 15;10:e13848. doi: 10.7717/peerj.13848. eCollection 2022. PeerJ. 2022. PMID: 35990901 Free PMC article.
-
Discovery of Potential Therapeutic Drugs for COVID-19 Through Logistic Matrix Factorization With Kernel Diffusion.Front Microbiol. 2022 Feb 28;13:740382. doi: 10.3389/fmicb.2022.740382. eCollection 2022. Front Microbiol. 2022. PMID: 35295301 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

