Genomes of Novel Myxococcota Reveal Severely Curtailed Machineries for Predation and Cellular Differentiation
- PMID: 34524899
- PMCID: PMC8580003
- DOI: 10.1128/AEM.01706-21
Genomes of Novel Myxococcota Reveal Severely Curtailed Machineries for Predation and Cellular Differentiation
Abstract
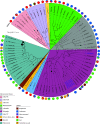
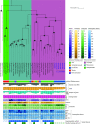
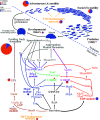
Cultured Myxococcota are predominantly aerobic soil inhabitants, characterized by their highly coordinated predation and cellular differentiation capacities. Little is currently known regarding yet-uncultured Myxococcota from anaerobic, nonsoil habitats. We analyzed genomes representing one novel order (o__JAFGXQ01) and one novel family (f__JAFGIB01) in the Myxococcota from an anoxic freshwater spring (Zodletone Spring) in Oklahoma, USA. Compared to their soil counterparts, anaerobic Myxococcota possess smaller genomes and a smaller number of genes encoding biosynthetic gene clusters (BGCs), peptidases, one- and two-component signal transduction systems, and transcriptional regulators. Detailed analysis of 13 distinct pathways/processes crucial to predation and cellular differentiation revealed severely curtailed machineries, with the notable absence of homologs for key transcription factors (e.g., FruA and MrpC), outer membrane exchange receptor (TraA), and the majority of sporulation-specific and A-motility-specific genes. Further, machine learning approaches based on a set of 634 genes informative of social lifestyle predicted a nonsocial behavior for Zodletone Myxococcota. Metabolically, Zodletone Myxococcota genomes lacked aerobic respiratory capacities but carried genes suggestive of fermentation, dissimilatory nitrite reduction, and dissimilatory sulfate-reduction (in f_JAFGIB01) for energy acquisition. We propose that predation and cellular differentiation represent a niche adaptation strategy that evolved circa 500 million years ago (Mya) in response to the rise of soil as a distinct habitat on Earth. IMPORTANCE The phylum Myxococcota is a phylogenetically coherent bacterial lineage that exhibits unique social traits. Cultured Myxococcota are predominantly aerobic soil-dwelling microorganisms that are capable of predation and fruiting body formation. However, multiple yet-uncultured lineages within the Myxococcota have been encountered in a wide range of nonsoil, predominantly anaerobic habitats, and the metabolic capabilities, physiological preferences, and capacity of social behavior of such lineages remain unclear. Here, we analyzed genomes recovered from a metagenomic analysis of an anoxic freshwater spring in Oklahoma, USA, that represent novel, yet-uncultured, orders and families in the Myxococcota. The genomes appear to lack the characteristic hallmarks for social behavior encountered in Myxococcota genomes and displayed a significantly smaller genome size and a smaller number of genes encoding biosynthetic gene clusters, peptidases, signal transduction systems, and transcriptional regulators. Such perceived lack of social capacity was confirmed through detailed comparative genomic analysis of 13 pathways associated with Myxococcota social behavior, as well as the implementation of machine learning approaches to predict social behavior based on genome composition. Metabolically, these novel Myxococcota are predicted to be strict anaerobes, utilizing fermentation, nitrate reduction, and dissimilarity sulfate reduction for energy acquisition. Our results highlight the broad patterns of metabolic diversity within the yet-uncultured Myxococcota and suggest that the evolution of predation and fruiting body formation in the Myxococcota has occurred in response to soil formation as a distinct habitat on Earth.
Keywords: Myxobacteria; fruiting body formation; genome resolved metagenomics; predation.
Figures
Similar articles
-
Genomic Analysis of Family UBA6911 (Group 18 Acidobacteria) Expands the Metabolic Capacities of the Phylum and Highlights Adaptations to Terrestrial Habitats.Appl Environ Microbiol. 2021 Aug 11;87(17):e0094721. doi: 10.1128/AEM.00947-21. Epub 2021 Aug 11. Appl Environ Microbiol. 2021. PMID: 34160232 Free PMC article.
-
Genomic Characterization of Candidate Division LCP-89 Reveals an Atypical Cell Wall Structure, Microcompartment Production, and Dual Respiratory and Fermentative Capacities.Appl Environ Microbiol. 2019 May 2;85(10):e00110-19. doi: 10.1128/AEM.00110-19. Print 2019 May 15. Appl Environ Microbiol. 2019. PMID: 30902854 Free PMC article.
-
Candidatus Krumholzibacterium zodletonense gen. nov., sp nov, the first representative of the candidate phylum Krumholzibacteriota phyl. nov. recovered from an anoxic sulfidic spring using genome resolved metagenomics.Syst Appl Microbiol. 2019 Jan;42(1):85-93. doi: 10.1016/j.syapm.2018.11.002. Epub 2018 Nov 15. Syst Appl Microbiol. 2019. PMID: 30477901
-
The genetic basis of predation by myxobacteria.Adv Microb Physiol. 2024;85:1-55. doi: 10.1016/bs.ampbs.2024.04.001. Epub 2024 May 22. Adv Microb Physiol. 2024. PMID: 39059819 Review.
-
Reduction of bacterial genome size and expansion resulting from obligate intracellular lifestyle and adaptation to soil habitat.Acta Biochim Pol. 2001;48(2):367-81. Acta Biochim Pol. 2001. PMID: 11732608 Review.
Cited by
-
Diverse secondary metabolites are expressed in particle-associated and free-living microorganisms of the permanently anoxic Cariaco Basin.Nat Commun. 2023 Feb 6;14(1):656. doi: 10.1038/s41467-023-36026-w. Nat Commun. 2023. PMID: 36746960 Free PMC article.
-
Characterization of endophytic bacteriome diversity and associated beneficial bacteria inhabiting a macrophyte Eichhornia crassipes.Front Plant Sci. 2023 Jun 19;14:1176648. doi: 10.3389/fpls.2023.1176648. eCollection 2023. Front Plant Sci. 2023. PMID: 37404529 Free PMC article.
-
The effects of adding exogenous lignocellulose degrading bacteria during straw incorporation in cold regions on degradation characteristics and soil indigenous bacteria communities.Front Microbiol. 2023 May 10;14:1141545. doi: 10.3389/fmicb.2023.1141545. eCollection 2023. Front Microbiol. 2023. PMID: 37234521 Free PMC article.
-
Novel candidate taxa contribute to key metabolic processes in Fennoscandian Shield deep groundwaters.ISME Commun. 2024 Sep 23;4(1):ycae113. doi: 10.1093/ismeco/ycae113. eCollection 2024 Jan. ISME Commun. 2024. PMID: 39421601 Free PMC article.
-
Supplementation of Dietary Crude Lentinan Improves the Intestinal Microbiota and Immune Barrier in Rainbow Trout (Oncorhynchus mykiss) Infected by Infectious Hematopoietic Necrosis Virus.Front Immunol. 2022 Jun 22;13:920065. doi: 10.3389/fimmu.2022.920065. eCollection 2022. Front Immunol. 2022. PMID: 35812417 Free PMC article.
References
-
- Shimkets LJ, Dworkin M, Reichenbach H. 2006. The Myxobacteria, p 31–115. In Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E (ed), The Prokaryotes, vol 7. Proteobacteria: delta, epsilon subclass. 10.1007/0-387-30747-8_3. Springer, New York, NY. - DOI
-
- Waite DW, Chuvochina M, Pelikan C, Parks DH, Yilmaz P, Wagner M, Loy A, Naganuma T, Nakai R, Whitman WB, Hahn MW, Kuever J, Hugenholtz P. 2020. Proposal to reclassify the proteobacterial classes Deltaproteobacteria and Oligoflexia, and the phylum Thermodesulfobacteria into four phyla reflecting major functional capabilities. Int J Syst Evol Microbiol 70:5972–6016. 10.1099/ijsem.0.004213. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

