CTCF as a regulator of alternative splicing: new tricks for an old player
- PMID: 34181707
- PMCID: PMC8373115
- DOI: 10.1093/nar/gkab520
CTCF as a regulator of alternative splicing: new tricks for an old player
Abstract
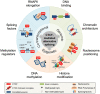
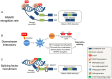
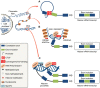
Three decades of research have established the CCCTC-binding factor (CTCF) as a ubiquitously expressed chromatin organizing factor and master regulator of gene expression. A new role for CTCF as a regulator of alternative splicing (AS) has now emerged. CTCF has been directly and indirectly linked to the modulation of AS at the individual transcript and at the transcriptome-wide level. The emerging role of CTCF-mediated regulation of AS involves diverse mechanisms; including transcriptional elongation, DNA methylation, chromatin architecture, histone modifications, and regulation of splicing factor expression and assembly. CTCF thereby appears to not only co-ordinate gene expression regulation but contributes to the modulation of transcriptomic complexity. In this review, we highlight previous discoveries regarding the role of CTCF in AS. In addition, we summarize detailed mechanisms by which CTCF mediates AS regulation. We propose opportunities for further research designed to examine the possible fate of CTCF-mediated alternatively spliced genes and associated biological consequences. CTCF has been widely acknowledged as the 'master weaver of the genome'. Given its multiple connections, further characterization of CTCF's emerging role in splicing regulation might extend its functional repertoire towards a 'conductor of the splicing orchestra'.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
Similar articles
-
CTCF-Mediated Chromatin Loops between Promoter and Gene Body Regulate Alternative Splicing across Individuals.Cell Syst. 2017 Dec 27;5(6):628-637.e6. doi: 10.1016/j.cels.2017.10.018. Epub 2017 Nov 29. Cell Syst. 2017. PMID: 29199022
-
Ctcf haploinsufficiency mediates intron retention in a tissue-specific manner.RNA Biol. 2021 Jan;18(1):93-103. doi: 10.1080/15476286.2020.1796052. Epub 2020 Aug 20. RNA Biol. 2021. PMID: 32816606 Free PMC article.
-
PARP1 Stabilizes CTCF Binding and Chromatin Structure To Maintain Epstein-Barr Virus Latency Type.J Virol. 2018 Aug 29;92(18):e00755-18. doi: 10.1128/JVI.00755-18. Print 2018 Sep 15. J Virol. 2018. PMID: 29976663 Free PMC article.
-
CTCF and Its Multi-Partner Network for Chromatin Regulation.Cells. 2023 May 10;12(10):1357. doi: 10.3390/cells12101357. Cells. 2023. PMID: 37408191 Free PMC article. Review.
-
CTCF: a Swiss-army knife for genome organization and transcription regulation.Essays Biochem. 2019 Apr 23;63(1):157-165. doi: 10.1042/EBC20180069. Print 2019 Apr 23. Essays Biochem. 2019. PMID: 30940740 Review.
Cited by
-
Cohesin-Mediated Chromatin Interactions and Autoimmunity.Front Immunol. 2022 Feb 10;13:840002. doi: 10.3389/fimmu.2022.840002. eCollection 2022. Front Immunol. 2022. PMID: 35222432 Free PMC article. Review.
-
Comprehensive analysis of pre-mRNA alternative splicing regulated by m6A methylation in pig oxidative and glycolytic skeletal muscles.BMC Genomics. 2022 Dec 6;23(1):804. doi: 10.1186/s12864-022-09043-0. BMC Genomics. 2022. PMID: 36474138 Free PMC article.
-
3C methods in cancer research: recent advances and future prospects.Exp Mol Med. 2024 Apr;56(4):788-798. doi: 10.1038/s12276-024-01236-9. Epub 2024 Apr 25. Exp Mol Med. 2024. PMID: 38658701 Free PMC article. Review.
-
CTCF: A misguided jack-of-all-trades in cancer cells.Comput Struct Biotechnol J. 2022 May 27;20:2685-2698. doi: 10.1016/j.csbj.2022.05.044. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 35685367 Free PMC article. Review.
-
3D chromatin architecture and transcription regulation in cancer.J Hematol Oncol. 2022 May 4;15(1):49. doi: 10.1186/s13045-022-01271-x. J Hematol Oncol. 2022. PMID: 35509102 Free PMC article. Review.
References
-
- Lobanenkov V.V., Nicolas R.H., Adler V.V., Paterson H., Klenova E.M., Polotskaja A.V., Goodwin G.H.. A novel sequence-specific DNA binding protein which interacts with three regularly spaced direct repeats of the CCCTC-motif in the 5′-flanking sequence of the chicken c-myc gene. Oncogene. 1990; 5:1743–1753. - PubMed
-
- Klenova E.M., Nicolas R.H., Paterson H.F., Carne A.F., Heath C.M., Goodwin G.H., Neiman P.E., Lobanenkov V.V.. CTCF, a conserved nuclear factor required for optimal transcriptional activity of the chicken c-myc gene, is an 11-Zn-finger protein differentially expressed in multiple forms. Mol. Cell. Biol. 1993; 13:7612–7624. - PMC - PubMed
-
- Bell A.C., West A.G., Felsenfeld G.. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999; 98:387–396. - PubMed
-
- Filippova G.N., Fagerlie S., Klenova E.M., Myers C., Dehner Y., Goodwin G., Neiman P.E., Collins S.J., Lobanenkov V.V.. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol. Cell. Biol. 1996; 16:2802–2813. - PMC - PubMed
-
- Ohlsson R., Renkawitz R., Lobanenkov V.. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet. 2001; 17:520–527. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

