The σ54 system directly regulates bacterial natural product genes
- PMID: 33637792
- PMCID: PMC7910581
- DOI: 10.1038/s41598-021-84057-4
The σ54 system directly regulates bacterial natural product genes
Abstract
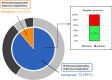
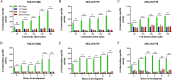
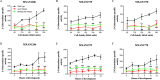
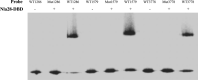
Bacterial-derived polyketide and non-ribosomal peptide natural products are crucial sources of therapeutics and yet little is known about the conditions that favor activation of natural product genes or the regulatory machinery controlling their transcription. Recent findings suggest that the σ54 system, which includes σ54-loaded RNA polymerase and transcriptional activators called enhancer binding proteins (EBPs), might be a common regulator of natural product genes. Here, we explored this idea by analyzing a selected group of putative σ54 promoters identified in Myxococcus xanthus natural product gene clusters. We show that mutations in putative σ54-RNA polymerase binding regions and in putative Nla28 EBP binding sites dramatically reduce in vivo promoter activities in growing and developing cells. We also show in vivo promoter activities are reduced in a nla28 mutant, that Nla28 binds to wild-type fragments of these promoters in vitro, and that in vitro binding is lost when the Nla28 binding sites are mutated. Together, our results indicate that M. xanthus uses σ54 promoters for transcription of at least some of its natural product genes. Interestingly, the vast majority of experimentally confirmed and putative σ54 promoters in M. xanthus natural product loci are located within genes and not in intergenic sequences.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Identifying the Gene Regulatory Network of the Starvation-Induced Transcriptional Activator Nla28.J Bacteriol. 2022 Dec 20;204(12):e0026522. doi: 10.1128/jb.00265-22. Epub 2022 Nov 30. J Bacteriol. 2022. PMID: 36448789 Free PMC article.
-
A cascade of coregulating enhancer binding proteins initiates and propagates a multicellular developmental program.Proc Natl Acad Sci U S A. 2011 Aug 9;108(32):E431-9. doi: 10.1073/pnas.1105876108. Epub 2011 Jun 13. Proc Natl Acad Sci U S A. 2011. PMID: 21670274 Free PMC article.
-
The Nla28S/Nla28 two-component signal transduction system regulates sporulation in Myxococcus xanthus.J Bacteriol. 2012 Sep;194(17):4698-708. doi: 10.1128/JB.00225-12. Epub 2012 Jun 29. J Bacteriol. 2012. PMID: 22753068 Free PMC article.
-
An early A-signal-dependent gene in Myxococcus xanthus has a sigma 54-like promoter.J Bacteriol. 1995 Aug;177(16):4638-44. doi: 10.1128/jb.177.16.4638-4644.1995. J Bacteriol. 1995. PMID: 7642489 Free PMC article.
-
The Regulatory Functions of σ54 Factor in Phytopathogenic Bacteria.Int J Mol Sci. 2021 Nov 24;22(23):12692. doi: 10.3390/ijms222312692. Int J Mol Sci. 2021. PMID: 34884502 Free PMC article. Review.
Cited by
-
Cell-cell transfer of adaptation traits benefits kin and actor in a cooperative microbe.Proc Natl Acad Sci U S A. 2024 Jul 23;121(30):e2402559121. doi: 10.1073/pnas.2402559121. Epub 2024 Jul 16. Proc Natl Acad Sci U S A. 2024. PMID: 39012831 Free PMC article.
-
Identifying the Gene Regulatory Network of the Starvation-Induced Transcriptional Activator Nla28.J Bacteriol. 2022 Dec 20;204(12):e0026522. doi: 10.1128/jb.00265-22. Epub 2022 Nov 30. J Bacteriol. 2022. PMID: 36448789 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

