Genetic Cell-Surface Modification for Optimized Foam Fractionation
- PMID: 33195133
- PMCID: PMC7658403
- DOI: 10.3389/fbioe.2020.572892
Genetic Cell-Surface Modification for Optimized Foam Fractionation
Abstract
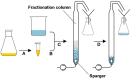
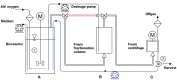
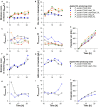
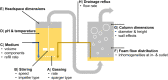
Rhamnolipids are among the glycolipids that have been investigated intensively in the last decades, mostly produced by the facultative pathogen Pseudomonas aeruginosa using plant oils as carbon source and antifoam agent. Simplification of downstream processing is envisaged using hydrophilic carbon sources, such as glucose, employing recombinant non-pathogenic Pseudomonas putida KT2440 for rhamnolipid or 3-(3-hydroxyalkanoyloxy)alkanoic acid (HAA, i.e., rhamnolipid precursors) production. However, during scale-up of the cultivation from shake flask to bioreactor, excessive foam formation hinders the use of standard fermentation protocols. In this study, the foam was guided from the reactor to a foam fractionation column to separate biosurfactants from medium and bacterial cells. Applying this integrated unit operation, the space-time yield (STY) for rhamnolipid synthesis could be increased by a factor of 2.8 (STY = 0.17 gRL/L·h) compared to the production in shake flasks. The accumulation of bacteria at the gas-liquid interface of the foam resulted in removal of whole-cell biocatalyst from the reactor with the strong consequence of reduced rhamnolipid production. To diminish the accumulation of bacteria at the gas-liquid interface, we deleted genes encoding cell-surface structures, focusing on hydrophobic proteins present on P. putida KT2440. Strains lacking, e.g., the flagellum, fimbriae, exopolysaccharides, and specific surface proteins, were tested for cell surface hydrophobicity and foam adsorption. Without flagellum or the large adhesion protein F (LapF), foam enrichment of these modified P. putida KT2440 was reduced by 23 and 51%, respectively. In a bioreactor cultivation of the non-motile strain with integrated rhamnolipid production genes, biomass enrichment in the foam was reduced by 46% compared to the reference strain. The intensification of rhamnolipid production from hydrophilic carbon sources presented here is an example for integrated strain and process engineering. This approach will become routine in the development of whole-cell catalysts for the envisaged bioeconomy. The results are discussed in the context of the importance of interacting strain and process engineering early in the development of bioprocesses.
Keywords: 3-(3-hydroxyalkanoyloxy)alkanoic acid (HAA); cell surface hydrophobicity; flagellum; foam fractionation; integrated product recovery; large adhesion protein; metabolic engineering; rhamnolipid.
Copyright © 2020 Blesken, Bator, Eberlein, Heipieper, Tiso and Blank.
Figures
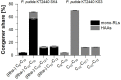



Similar articles
-
Uncoupling Foam Fractionation and Foam Adsorption for Enhanced Biosurfactant Synthesis and Recovery.Microorganisms. 2020 Dec 18;8(12):2029. doi: 10.3390/microorganisms8122029. Microorganisms. 2020. PMID: 33353027 Free PMC article.
-
Growth independent rhamnolipid production from glucose using the non-pathogenic Pseudomonas putida KT2440.Microb Cell Fact. 2011 Oct 17;10:80. doi: 10.1186/1475-2859-10-80. Microb Cell Fact. 2011. PMID: 21999513 Free PMC article.
-
Integrated foam fractionation for heterologous rhamnolipid production with recombinant Pseudomonas putida in a bioreactor.AMB Express. 2016 Mar;6(1):11. doi: 10.1186/s13568-016-0183-2. Epub 2016 Feb 9. AMB Express. 2016. PMID: 26860613 Free PMC article.
-
Rhamnolipids--next generation surfactants?J Biotechnol. 2012 Dec 31;162(4):366-80. doi: 10.1016/j.jbiotec.2012.05.022. Epub 2012 Jun 20. J Biotechnol. 2012. PMID: 22728388 Review.
-
Regulatory and metabolic network of rhamnolipid biosynthesis: traditional and advanced engineering towards biotechnological production.Appl Microbiol Biotechnol. 2011 Jul;91(2):251-64. doi: 10.1007/s00253-011-3368-2. Epub 2011 Jun 11. Appl Microbiol Biotechnol. 2011. PMID: 21667084 Review.
Cited by
-
Integration of Genetic and Process Engineering for Optimized Rhamnolipid Production Using Pseudomonas putida.Front Bioeng Biotechnol. 2020 Aug 20;8:976. doi: 10.3389/fbioe.2020.00976. eCollection 2020. Front Bioeng Biotechnol. 2020. PMID: 32974309 Free PMC article.
-
Identification and quantification of biosurfactants produced by the marine bacterium Alcanivorax borkumensis by hyphenated techniques.Anal Bioanal Chem. 2023 Dec;415(29-30):7067-7084. doi: 10.1007/s00216-023-04972-5. Epub 2023 Oct 11. Anal Bioanal Chem. 2023. PMID: 37819435 Free PMC article.
-
A Straightforward Assay for Screening and Quantification of Biosurfactants in Microbial Culture Supernatants.Front Bioeng Biotechnol. 2020 Aug 20;8:958. doi: 10.3389/fbioe.2020.00958. eCollection 2020. Front Bioeng Biotechnol. 2020. PMID: 32974305 Free PMC article.
-
Coupling an Electroactive Pseudomonas putida KT2440 with Bioelectrochemical Rhamnolipid Production.Microorganisms. 2020 Dec 10;8(12):1959. doi: 10.3390/microorganisms8121959. Microorganisms. 2020. PMID: 33322018 Free PMC article.
-
Uncoupling Foam Fractionation and Foam Adsorption for Enhanced Biosurfactant Synthesis and Recovery.Microorganisms. 2020 Dec 18;8(12):2029. doi: 10.3390/microorganisms8122029. Microorganisms. 2020. PMID: 33353027 Free PMC article.
References
-
- Bagdasarian M., Lurz R., Rückert B., Franklin F. C. H., Bagdasarian M. M., Frey J., et al. . (1981). Specific-purpose plasmid cloning vectors II. Broad host range, high copy number, RSF 1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene 16, 237-247. 10.1016/0378-1119(81)90080-9 - DOI - PubMed
LinkOut - more resources
Full Text Sources

