AmrZ and FleQ Co-regulate Cellulose Production in Pseudomonas syringae pv. Tomato DC3000
- PMID: 31057500
- PMCID: PMC6478803
- DOI: 10.3389/fmicb.2019.00746
AmrZ and FleQ Co-regulate Cellulose Production in Pseudomonas syringae pv. Tomato DC3000
Abstract
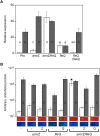
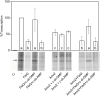
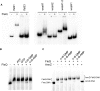
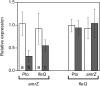
Pseudomonas syringae pv. tomato DC3000 carries the wssABCDEFGHI operon for the synthesis of acetylated cellulose, whose production is stimulated by increasing the intracellular levels of the second messenger c-di-GMP. This enhances air-liquid biofilm formation and generates a wrinkly colony morphotype in solid media. In the present study we show that cellulose production is a complex process regulated at multiple levels and involving different players in this bacterium. Using different in vitro approaches, including Electrophoretic Mobility Shift Assay (EMSA) and footprint analysis, we demonstrated the interrelated role of two transcriptional regulators, AmrZ and FleQ, over cellulose production in Pto DC3000 and the influence of c-di-GMP in this process. Under physiological c-di-GMP levels, both regulators bind directly to adjacent regions at the wss promoter inhibiting its expression. However, just FleQ responds to c-di-GMP releasing from its wss operator site and converting from a repressor to an activator of cellulose production. The additive effect of the double amrZ/fleQ mutation on the expression of wss, together with the fact that they are not cross-regulated at the transcriptional level, suggest that FleQ and AmrZ behave as independent regulators, unlike what has been described in other Pseudomonas species. Furthermore, this dual co-regulation exerted by AmrZ and FleQ is not limited to cellulose production, but also affects other important phenotypes in Pto DC3000, such as motility and virulence.
Keywords: AmrZ; FleQ; Pseudomonas syringae; c-di-GMP; cellulose; transcriptional regulation.
Figures


Similar articles
-
FleQ, FleN and c-di-GMP coordinately regulate cellulose production in Pseudomonas syringae pv. tomato DC3000.Front Mol Biosci. 2023 Mar 27;10:1155579. doi: 10.3389/fmolb.2023.1155579. eCollection 2023. Front Mol Biosci. 2023. PMID: 37051327 Free PMC article.
-
AmrZ regulates cellulose production in Pseudomonas syringae pv. tomato DC3000.Mol Microbiol. 2016 Mar;99(5):960-77. doi: 10.1111/mmi.13278. Epub 2015 Dec 15. Mol Microbiol. 2016. PMID: 26564578
-
Visualization and characterization of Pseudomonas syringae pv. tomato DC3000 pellicles.Microb Biotechnol. 2019 Jul;12(4):688-702. doi: 10.1111/1751-7915.13385. Epub 2019 Mar 5. Microb Biotechnol. 2019. PMID: 30838765 Free PMC article.
-
Adaption of Pseudomonas ogarae F113 to the Rhizosphere Environment-The AmrZ-FleQ Hub.Microorganisms. 2023 Apr 15;11(4):1037. doi: 10.3390/microorganisms11041037. Microorganisms. 2023. PMID: 37110460 Free PMC article. Review.
-
Flip the switch: the role of FleQ in modulating the transition between the free-living and sessile mode of growth in Pseudomonas aeruginosa.J Bacteriol. 2024 Mar 21;206(3):e0036523. doi: 10.1128/jb.00365-23. Epub 2024 Mar 4. J Bacteriol. 2024. PMID: 38436566 Free PMC article. Review.
Cited by
-
Transcriptomic analysis of Pseudomonas ogarae F113 reveals the antagonistic roles of AmrZ and FleQ during rhizosphere adaption.Microb Genom. 2022 Jan;8(1):000750. doi: 10.1099/mgen.0.000750. Microb Genom. 2022. PMID: 35012704 Free PMC article.
-
Quantification of Mixed-Linkage β-Glucan (MLG) in Bacteria.Methods Mol Biol. 2024;2751:133-143. doi: 10.1007/978-1-0716-3617-6_9. Methods Mol Biol. 2024. PMID: 38265714
-
Biological role of EPS from Pseudomonas syringae pv. syringae UMAF0158 extracellular matrix, focusing on a Psl-like polysaccharide.NPJ Biofilms Microbiomes. 2020 Oct 12;6(1):37. doi: 10.1038/s41522-020-00148-6. NPJ Biofilms Microbiomes. 2020. PMID: 33046713 Free PMC article.
-
Pervasive RNA Regulation of Metabolism Enhances the Root Colonization Ability of Nitrogen-Fixing Symbiotic α-Rhizobia.mBio. 2021 Feb 22;13(1):e0357621. doi: 10.1128/mbio.03576-21. Epub 2022 Feb 15. mBio. 2021. PMID: 35164560 Free PMC article.
-
Pseudomonas syringae addresses distinct environmental challenges during plant infection through the coordinated deployment of polysaccharides.J Exp Bot. 2022 Apr 5;73(7):2206-2221. doi: 10.1093/jxb/erab550. J Exp Bot. 2022. PMID: 34905021 Free PMC article.
References
-
- Baraquet C., Murakami K., Parsek M. R., Harwood C. S. (2012). The FleQ protein from Pseudomonas aeruginosa functions as both a repressor and an activator to control gene expression from the pel operon promoter in response to c-di-GMP. Nucleic Acids Res. 40 7207–7218. 10.1093/nar/gks384 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

