Suppressor mutations reveal an NtrC-like response regulator, NmpR, for modulation of Type-IV Pili-dependent motility in Myxococcus xanthus
- PMID: 30346960
- PMCID: PMC6211767
- DOI: 10.1371/journal.pgen.1007714
Suppressor mutations reveal an NtrC-like response regulator, NmpR, for modulation of Type-IV Pili-dependent motility in Myxococcus xanthus
Abstract
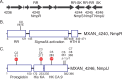
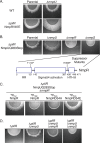
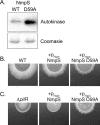
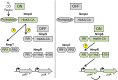
Two-component signaling systems (TCS) regulate bacterial responses to environmental signals through the process of protein phosphorylation. Specifically, sensor histidine kinases (SK) recognize signals and propagate the response via phosphorylation of a cognate response regulator (RR) that functions to initiate transcription of specific genes. Signaling within a single TCS is remarkably specific and cross-talk between TCS is limited. However, regulation of the flow of information through complex signaling networks that include closely related TCS remains largely unknown. Additionally, many bacteria utilize multi-component signaling networks which provide additional genetic and biochemical interactions that must be regulated for signaling fidelity, input and output specificity, and phosphorylation kinetics. Here we describe the characterization of an NtrC-like RR that participates in regulation of Type-IV pilus-dependent motility of Myxococcus xanthus and is thus named NmpR, NtrC Modulator of Pili Regulator. A complex multi-component signaling system including NmpR was revealed by suppressor mutations that restored motility to cells lacking PilR, an evolutionarily conserved RR required for expression of pilA encoding the major Type-IV pilus monomer found in many bacterial species. The system contains at least four signaling proteins: a SK with a protoglobin sensor domain (NmpU), a hybrid SK (NmpS), a phospho-sink protein (NmpT), and an NtrC-like RR (NmpR). We demonstrate that ΔpilR bypass suppressor mutations affect regulation of the NmpRSTU multi-component system, such that NmpR activation is capable of restoring expression of pilA in the absence of PilR. Our findings indicate that pilus gene expression in M. xanthus is regulated by an extended network of TCS which interact to refine control of pilus function.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





Similar articles
-
The Pseudomonas aeruginosa PilSR Two-Component System Regulates Both Twitching and Swimming Motilities.mBio. 2018 Jul 24;9(4):e01310-18. doi: 10.1128/mBio.01310-18. mBio. 2018. PMID: 30042200 Free PMC article.
-
Type IV-pili dependent motility is co-regulated by PilSR and PilS2R2 two-component systems via distinct pathways in Myxococcus xanthus.Mol Microbiol. 2016 Oct;102(1):37-53. doi: 10.1111/mmi.13445. Epub 2016 Aug 23. Mol Microbiol. 2016. PMID: 27393239
-
The Myxococcus xanthus pilT locus is required for social gliding motility although pili are still produced.Mol Microbiol. 1997 Jan;23(1):109-21. doi: 10.1046/j.1365-2958.1997.1791550.x. Mol Microbiol. 1997. PMID: 9004225
-
Dual regulation with Ser/Thr kinase cascade and a His/Asp TCS in Myxococcus xanthus.Adv Exp Med Biol. 2008;631:111-21. doi: 10.1007/978-0-387-78885-2_7. Adv Exp Med Biol. 2008. PMID: 18792684 Review.
-
The type IVc pilus: just a Tad different.Curr Opin Microbiol. 2024 Jun;79:102468. doi: 10.1016/j.mib.2024.102468. Epub 2024 Apr 4. Curr Opin Microbiol. 2024. PMID: 38579360 Review.
Cited by
-
A miniTurbo-based proximity labeling protocol to identify conditional protein interactomes in vivo in Myxococcus xanthus.STAR Protoc. 2023 Dec 15;4(4):102657. doi: 10.1016/j.xpro.2023.102657. Epub 2023 Oct 25. STAR Protoc. 2023. PMID: 37883223 Free PMC article.
-
Characterization of a Cobalt-Substituted Globin-Coupled Oxygen Sensor Histidine Kinase from Anaeromyxobacter sp. Fw109-5: Insights into Catalytic Regulation by Its Heme Coordination Structure.ACS Omega. 2021 Dec 6;6(50):34912-34919. doi: 10.1021/acsomega.1c05564. eCollection 2021 Dec 21. ACS Omega. 2021. PMID: 34963974 Free PMC article.
-
Evolutionary innovation through transcription factor rewiring in microbes is shaped by levels of transcription factor activity, expression, and existing connectivity.PLoS Biol. 2023 Oct 23;21(10):e3002348. doi: 10.1371/journal.pbio.3002348. eCollection 2023 Oct. PLoS Biol. 2023. PMID: 37871011 Free PMC article.
-
Cell-cell transfer of adaptation traits benefits kin and actor in a cooperative microbe.Proc Natl Acad Sci U S A. 2024 Jul 23;121(30):e2402559121. doi: 10.1073/pnas.2402559121. Epub 2024 Jul 16. Proc Natl Acad Sci U S A. 2024. PMID: 39012831 Free PMC article.
References
-
- Hobbs M, Collie ES, Free PD, Livingston SP, Mattick JS. PilS and PilR, a two-component transcriptional regulatory system controlling expression of type 4 fimbriae in Pseudomonas aeruginosa. Mol Microbiol. 1993;7(5):669–82. . - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

