Making and Breaking of an Essential Poison: the Cyclases and Phosphodiesterases That Produce and Degrade the Essential Second Messenger Cyclic di-AMP in Bacteria
- PMID: 30224435
- PMCID: PMC6287462
- DOI: 10.1128/JB.00462-18
Making and Breaking of an Essential Poison: the Cyclases and Phosphodiesterases That Produce and Degrade the Essential Second Messenger Cyclic di-AMP in Bacteria
Abstract
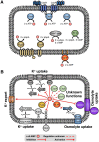
Cyclic di-AMP is a second-messenger nucleotide that is produced by many bacteria and some archaea. Recent work has shown that c-di-AMP is unique among the signaling nucleotides, as this molecule is in many bacteria both essential on one hand and toxic upon accumulation on the other. Moreover, in bacteria, like Bacillus subtilis, c-di-AMP controls a biological process, potassium homeostasis, by binding both potassium transporters and riboswitch molecules in the mRNAs that encode the potassium transporters. In addition to the control of potassium homeostasis, c-di-AMP has been implicated in many cellular activities, including DNA repair, cell wall homeostasis, osmotic adaptation, biofilm formation, central metabolism, and virulence. c-di-AMP is synthesized and degraded by diadenylate cyclases and phosphodiesterases, respectively. In the diadenylate cyclases, one type of catalytic domain, the diadenylate cyclase (DAC) domain, is coupled to various other domains that control the localization, the protein-protein interactions, and the regulation of the enzymes. The phosphodiesterases have a catalytic core that consists either of a DHH/DHHA1 or of an HD domain. Recent findings on the occurrence, domain organization, activity control, and structural features of diadenylate cyclases and phosphodiesterases are discussed in this review.
Keywords: diadenylate cyclase; phosphodiesterase; second messenger; signal transduction.
Copyright © 2018 American Society for Microbiology.
Figures


Similar articles
-
An Essential Poison: Synthesis and Degradation of Cyclic Di-AMP in Bacillus subtilis.J Bacteriol. 2015 Oct;197(20):3265-74. doi: 10.1128/JB.00564-15. Epub 2015 Aug 3. J Bacteriol. 2015. PMID: 26240071 Free PMC article.
-
A jack of all trades: the multiple roles of the unique essential second messenger cyclic di-AMP.Mol Microbiol. 2015 Jul;97(2):189-204. doi: 10.1111/mmi.13026. Epub 2015 May 9. Mol Microbiol. 2015. PMID: 25869574 Review.
-
Functional Analysis of a c-di-AMP-specific Phosphodiesterase MsPDE from Mycobacterium smegmatis.Int J Biol Sci. 2015 May 30;11(7):813-24. doi: 10.7150/ijbs.11797. eCollection 2015. Int J Biol Sci. 2015. PMID: 26078723 Free PMC article.
-
All DACs in a Row: Domain Architectures of Bacterial and Archaeal Diadenylate Cyclases.J Bacteriol. 2023 Apr 25;205(4):e0002323. doi: 10.1128/jb.00023-23. Epub 2023 Apr 6. J Bacteriol. 2023. PMID: 37022175 Free PMC article. Review.
-
YybT is a signaling protein that contains a cyclic dinucleotide phosphodiesterase domain and a GGDEF domain with ATPase activity.J Biol Chem. 2010 Jan 1;285(1):473-82. doi: 10.1074/jbc.M109.040238. Epub 2009 Nov 9. J Biol Chem. 2010. PMID: 19901023 Free PMC article.
Cited by
-
Nano-RNases: oligo- or dinucleases?FEMS Microbiol Rev. 2022 Nov 2;46(6):fuac038. doi: 10.1093/femsre/fuac038. FEMS Microbiol Rev. 2022. PMID: 36026528 Free PMC article. Review.
-
Borderline resistance to oxacillin in Staphylococcus aureus after treatment with sub-lethal sodium hypochlorite concentrations.Heliyon. 2020 Jun 21;6(6):e04070. doi: 10.1016/j.heliyon.2020.e04070. eCollection 2020 Jun. Heliyon. 2020. PMID: 32613099 Free PMC article.
-
An emerging role for cyclic dinucleotide phosphodiesterase and nanoRNase activities in Mycoplasma bovis: Securing survival in cell culture.PLoS Pathog. 2020 Jun 29;16(6):e1008661. doi: 10.1371/journal.ppat.1008661. eCollection 2020 Jun. PLoS Pathog. 2020. PMID: 32598377 Free PMC article.
-
Identification of the main glutamine and glutamate transporters in Staphylococcus aureus and their impact on c-di-AMP production.Mol Microbiol. 2020 Jun;113(6):1085-1100. doi: 10.1111/mmi.14479. Epub 2020 Feb 11. Mol Microbiol. 2020. PMID: 31997474 Free PMC article.
-
Fluorometric Liposome Screen for Inhibitors of a Physiologically Important Bacterial Ion Channel.Front Microbiol. 2021 Mar 1;12:603700. doi: 10.3389/fmicb.2021.603700. eCollection 2021. Front Microbiol. 2021. PMID: 33732218 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

