Mutation update of transcription factor genes FOXE3, HSF4, MAF, and PITX3 causing cataracts and other developmental ocular defects
- PMID: 29314435
- PMCID: PMC5839989
- DOI: 10.1002/humu.23395
Mutation update of transcription factor genes FOXE3, HSF4, MAF, and PITX3 causing cataracts and other developmental ocular defects
Abstract
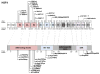
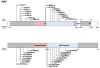
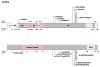
Mutations in the transcription factor genes FOXE3, HSF4, MAF, and PITX3 cause congenital lens defects including cataracts that may be accompanied by defects in other components of the eye or in nonocular tissues. We comprehensively describe here all the variants in FOXE3, HSF4, MAF, and PITX3 genes linked to human developmental defects. A total of 52 variants for FOXE3, 18 variants for HSF4, 20 variants for MAF, and 19 variants for PITX3 identified so far in isolated cases or within families are documented. This effort reveals FOXE3, HSF4, MAF, and PITX3 to have 33, 16, 18, and 7 unique causal mutations, respectively. Loss-of-function mutant animals for these genes have served to model the pathobiology of the associated human defects, and we discuss the currently known molecular function of these genes, particularly with emphasis on their role in ocular development. Finally, we make the detailed FOXE3, HSF4, MAF, and PITX3 variant information available in the Leiden Online Variation Database (LOVD) platform at https://www.LOVD.nl/FOXE3, https://www.LOVD.nl/HSF4, https://www.LOVD.nl/MAF, and https://www.LOVD.nl/PITX3. Thus, this article informs on key variants in transcription factor genes linked to cataract, aphakia, corneal opacity, glaucoma, microcornea, microphthalmia, anterior segment mesenchymal dysgenesis, and Ayme-Gripp syndrome, and facilitates their access through Web-based databases.
Keywords: Ayme-Gripp syndrome; LOVD; anterior segment mesenchymal dysgenesis; aphakia; cataract; microcornea.
© 2018 Wiley Periodicals, Inc.
Conflict of interest statement
Figures





Similar articles
-
Ayme gripp syndrome in an Indian patient.Am J Med Genet A. 2021 Apr;185(4):1312-1316. doi: 10.1002/ajmg.a.62053. Epub 2021 Jan 1. Am J Med Genet A. 2021. PMID: 33528093
-
Skeletal abnormalities are common features in Aymé-Gripp syndrome.Clin Genet. 2020 Feb;97(2):362-369. doi: 10.1111/cge.13651. Epub 2019 Nov 3. Clin Genet. 2020. PMID: 31600839
-
Further delineation of Aymé-Gripp syndrome and use of automated facial analysis tool.Am J Med Genet A. 2018 Jul;176(7):1648-1656. doi: 10.1002/ajmg.a.38832. Am J Med Genet A. 2018. PMID: 30160832
-
Identification of a novel missense mutation of MAF in a Japanese family with congenital cataract by whole exome sequencing: a clinical report and review of literature.Am J Med Genet A. 2014 May;164A(5):1272-6. doi: 10.1002/ajmg.a.36433. Epub 2014 Mar 24. Am J Med Genet A. 2014. PMID: 24664492 Review.
-
FOXE3 mutations: genotype-phenotype correlations.Clin Genet. 2018 Apr;93(4):837-845. doi: 10.1111/cge.13177. Clin Genet. 2018. PMID: 29136273 Review.
Cited by
-
Mapping the Universe of Eph Receptor and Ephrin Ligand Transcripts in Epithelial and Fiber Cells of the Eye Lens.Cells. 2022 Oct 19;11(20):3291. doi: 10.3390/cells11203291. Cells. 2022. PMID: 36291158 Free PMC article.
-
Congenital cataract: a guide to genetic and clinical management.Ther Adv Rare Dis. 2020 Jul 22;1:2633004020938061. doi: 10.1177/2633004020938061. eCollection 2020 Jan-Dec. Ther Adv Rare Dis. 2020. PMID: 37180497 Free PMC article. Review.
-
Molecular and Genetic Mechanism of Non-Syndromic Congenital Cataracts. Mutation Screening in Spanish Families.Genes (Basel). 2021 Apr 16;12(4):580. doi: 10.3390/genes12040580. Genes (Basel). 2021. PMID: 33923544 Free PMC article.
-
Deficiency of the bZIP transcription factors Mafg and Mafk causes misexpression of genes in distinct pathways and results in lens embryonic developmental defects.Front Cell Dev Biol. 2022 Aug 26;10:981893. doi: 10.3389/fcell.2022.981893. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36092713 Free PMC article.
-
Stimulation of C-Kit+ Retinal Progenitor Cells by Stem Cell Factor Confers Protection Against Retinal Degeneration.Front Pharmacol. 2022 Mar 31;13:796380. doi: 10.3389/fphar.2022.796380. eCollection 2022. Front Pharmacol. 2022. PMID: 35431956 Free PMC article.
References
-
- Agrawal SA, Anand D, Siddam AD, Kakrana A, Dash S, Scheiblin DA, Dang CA, Terrell AM, Waters SM, Singh A, Motohashi H, Yamamoto M, et al. Compound mouse mutants of bZIP transcription factors Mafg and Mafk reveal a regulatory network of non-crystallin genes associated with cataract. Hum Genet. 2015;134:717–735. - PMC - PubMed
-
- Ahmad N, Aslam M, Muenster D, Horsch M, Khan MA, Carlsson P, Beckers J, Graw J. Pitx3 directly regulates Foxe3 during early lens development. Int J Dev Biol. 2013;57:741–751. - PubMed
-
- Aldahmesh MA, Khan AO, Mohamed J, Alkuraya FS. Novel recessive BFSP2 and PITX3 mutations: insights into mutational mechanisms from consanguineous populations. Genet Med Off J Am Coll Med Genet. 2011;13:978–981. - PubMed
-
- Ali M, Buentello-Volante B, McKibbin M, Rocha-Medina JA, Fernandez-Fuentes N, Koga-Nakamura W, Ashiq A, Khan K, Booth AP, Williams G, Raashid Y, Jafri H, et al. Homozygous FOXE3 mutations cause non-syndromic, bilateral, total sclerocornea, aphakia, microphthalmia and optic disc coloboma. Mol Vis. 2010;16:1162–1168. - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

