Initial Cluster Analysis
- PMID: 28771374
- PMCID: PMC5806593
- DOI: 10.1089/cmb.2017.0050
Initial Cluster Analysis
Abstract
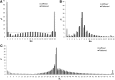
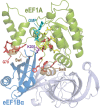
We study a simple abstract problem motivated by a variety of applications in protein sequence analysis. Consider a string of 0s and 1s of length L, and containing D 1s. If we believe that some or all of the 1s may be clustered near the start of the sequence, which subset is the most significantly so clustered, and how significant is this clustering? We approach this question using the minimum description length principle and illustrate its application by analyzing residues that distinguish translational initiation and elongation factor guanosine triphosphatases (GTPases) from other P-loop GTPases. Within a structure of yeast elongation factor 1[Formula: see text], these residues form a significant cluster centered on a region implicated in guanine nucleotide exchange. Various biomedical questions may be cast as the abstract problem considered here.
Keywords: Jeffreys' priors; Minimum Description Length principle; cluster analysis.
Conflict of interest statement
The authors declare that there are no competing financial interests.
Figures

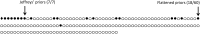
Similar articles
-
Classification and evolution of P-loop GTPases and related ATPases.J Mol Biol. 2002 Mar 15;317(1):41-72. doi: 10.1006/jmbi.2001.5378. J Mol Biol. 2002. PMID: 11916378
-
Translation elongation factor-3 (EF-3): an evolving eukaryotic ribosomal protein?J Mol Evol. 1995 Sep;41(3):376-87. J Mol Evol. 1995. PMID: 7563124
-
[GTPases of translational apparatus].Mol Biol (Mosk). 2005 Sep-Oct;39(5):746-61. Mol Biol (Mosk). 2005. PMID: 16240709 Review. Russian.
-
Ran's C-terminal, basic patch, and nucleotide exchange mechanisms in light of a canonical structure for Rab, Rho, Ras, and Ran GTPases.Genome Res. 2003 Apr;13(4):673-92. doi: 10.1101/gr.862303. Genome Res. 2003. PMID: 12671004 Free PMC article.
-
Clustering algorithms in biomedical research: a review.IEEE Rev Biomed Eng. 2010;3:120-54. doi: 10.1109/RBME.2010.2083647. IEEE Rev Biomed Eng. 2010. PMID: 22275205 Review.
Cited by
-
SPARC: Structural properties associated with residue constraints.Comput Struct Biotechnol J. 2022 Apr 7;20:1702-1715. doi: 10.1016/j.csbj.2022.04.005. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 35495120 Free PMC article.
-
Inferring joint sequence-structural determinants of protein functional specificity.Elife. 2018 Jan 16;7:e29880. doi: 10.7554/eLife.29880. Elife. 2018. PMID: 29336305 Free PMC article.
-
Identifying Function Determining Residues in Neuroimmune Semaphorin 4A.Int J Mol Sci. 2022 Mar 11;23(6):3024. doi: 10.3390/ijms23063024. Int J Mol Sci. 2022. PMID: 35328445 Free PMC article.
-
Statistical investigations of protein residue direct couplings.PLoS Comput Biol. 2018 Dec 31;14(12):e1006237. doi: 10.1371/journal.pcbi.1006237. eCollection 2018 Dec. PLoS Comput Biol. 2018. PMID: 30596639 Free PMC article.
References
-
-
Andersen G.R., Valente L., Pedersen L., et al. . 2001. Crystal structures of nucleotide exchange intermediates in the eEF0A-eEF1B
complex. Nat. Struct. Biol. 8, 531–534 - PubMed
-
Andersen G.R., Valente L., Pedersen L., et al. . 2001. Crystal structures of nucleotide exchange intermediates in the eEF0A-eEF1B
-
- Durbin R., Eddy S., Krogh A., et al. . 1998. Biological Sequence Analysis. Probabilistic Models of Proteins and Nucleic Acids. Cambridge University Press, Cambridge, England
-
- Fischer J.D., E., Mayer C., and Söding J. 2008. Prediction of protein functional residues from sequence by probability density estimation. Bioinformatics 24, 613–620 - PubMed
-
- Grünwald P.D. 2007. The Minimum Description Length Principle. MIT Press, Cambridge, MA
-
- Hall A. 2000. GTPases. Oxford University Press, Oxford, England
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

