IBD Sharing between Africans, Neandertals, and Denisovans
- PMID: 28158547
- PMCID: PMC5381509
- DOI: 10.1093/gbe/evw234
IBD Sharing between Africans, Neandertals, and Denisovans
Abstract
Interbreeding between ancestors of humans and other hominins outside of Africa has been studied intensively, while their common history within Africa still lacks proper attention. However, shedding light on human evolution in this time period about which little is known, is essential for understanding subsequent events outside of Africa. We investigate the genetic relationships of humans, Neandertals, and Denisovans by identifying very short DNA segments in the 1000 Genomes Phase 3 data that these hominins share identical by descent (IBD). By focusing on low frequency and rare variants, we identify very short IBD segments with high confidence. These segments reveal events from a very distant past because shorter IBD segments are presumably older than longer ones. We extracted two types of very old IBD segments that are not only shared among humans, but also with Neandertals and/or Denisovans. The first type contains longer segments that are found primarily in Asians and Europeans where more segments are found in South Asians than in East Asians for both Neandertal and Denisovan. These longer segments indicate complex admixture events outside of Africa. The second type consists of shorter segments that are shared mainly by Africans and therefore may indicate events involving ancestors of humans and other ancient hominins within Africa. Our results from the autosomes are further supported by an analysis of chromosome X, on which segments that are shared by Africans and match the Neandertal and/or Denisovan genome were even more prominent. Our results indicate that interbreeding with other hominins was a common feature of human evolution starting already long before ancestors of modern humans left Africa.
Keywords: identity by descent; interbreeding; Denisova; human evolution; gene flow; Neandertal.
Figures
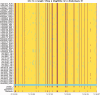
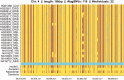
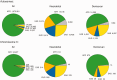
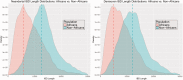
Similar articles
-
The date of interbreeding between Neandertals and modern humans.PLoS Genet. 2012;8(10):e1002947. doi: 10.1371/journal.pgen.1002947. Epub 2012 Oct 4. PLoS Genet. 2012. PMID: 23055938 Free PMC article.
-
Detecting archaic introgression using an unadmixed outgroup.PLoS Genet. 2018 Sep 18;14(9):e1007641. doi: 10.1371/journal.pgen.1007641. eCollection 2018 Sep. PLoS Genet. 2018. PMID: 30226838 Free PMC article.
-
Complex history of admixture between modern humans and Neandertals.Am J Hum Genet. 2015 Mar 5;96(3):448-53. doi: 10.1016/j.ajhg.2015.01.006. Epub 2015 Feb 12. Am J Hum Genet. 2015. PMID: 25683119 Free PMC article.
-
Archaic hominin introgression into modern human genomes.Am J Phys Anthropol. 2020 May;171 Suppl 70:60-73. doi: 10.1002/ajpa.23951. Epub 2019 Nov 8. Am J Phys Anthropol. 2020. PMID: 31702050 Review.
-
Ancient humans and the origin of modern humans.Curr Opin Genet Dev. 2014 Dec;29:133-8. doi: 10.1016/j.gde.2014.09.004. Epub 2014 Oct 3. Curr Opin Genet Dev. 2014. PMID: 25286439 Review.
Cited by
-
Temporal population structure, a genetic dating method for ancient Eurasian genomes from the past 10,000 years.Cell Rep Methods. 2022 Aug 22;2(8):100270. doi: 10.1016/j.crmeth.2022.100270. eCollection 2022 Aug 22. Cell Rep Methods. 2022. PMID: 36046618 Free PMC article.
-
Multiple selective sweeps of ancient polymorphisms in and around LTα located in the MHC class III region on chromosome 6.BMC Evol Biol. 2019 Dec 2;19(1):218. doi: 10.1186/s12862-019-1516-y. BMC Evol Biol. 2019. PMID: 31791241 Free PMC article.
-
Global Picture of Genetic Relatedness and the Evolution of Humankind.Biology (Basel). 2020 Nov 10;9(11):392. doi: 10.3390/biology9110392. Biology (Basel). 2020. PMID: 33182715 Free PMC article.
-
Rectified factor networks for biclustering of omics data.Bioinformatics. 2017 Jul 15;33(14):i59-i66. doi: 10.1093/bioinformatics/btx226. Bioinformatics. 2017. PMID: 28881961 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

