Signal Transducer and Activator of Transcription 1 Plays a Pivotal Role in RET/PTC3 Oncogene-induced Expression of Indoleamine 2,3-Dioxygenase 1
- PMID: 27994058
- PMCID: PMC5290952
- DOI: 10.1074/jbc.M116.745448
Signal Transducer and Activator of Transcription 1 Plays a Pivotal Role in RET/PTC3 Oncogene-induced Expression of Indoleamine 2,3-Dioxygenase 1
Abstract
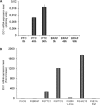
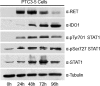
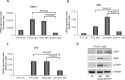
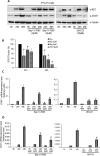
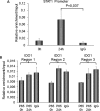
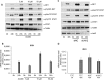
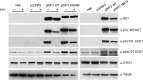
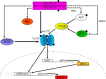
Indoleamine 2,3-dioxygenase 1 (IDO1) is a single chain oxidoreductase that catalyzes tryptophan degradation to kynurenine. In cancer, it exerts an immunosuppressive function as part of an acquired mechanism of immune escape. Recently, we demonstrated that IDO1 expression is significantly higher in all thyroid cancer histotypes compared with normal thyroid and that its expression levels correlate with T regulatory (Treg) lymphocyte densities in the tumor microenvironment. BRAFV600E- and RET/PTC3-expressing PcCL3 cells were used as cellular models for the evaluation of IDO1 expression in thyroid carcinoma cells and for the study of involved signal transduction pathways. BRAFV600E-expressing PcCL3 cells did not show IDO1 expression. Conversely, RET/PTC3-expressing cells were characterized by a high IDO1 expression. Moreover, we found that, the STAT1-IRF1 pathway was instrumental for IDO1 expression in RET/PTC3 expressing cells. In detail, RET/PTC3 induced STAT1 overexpression and phosphorylation at Ser-727 and Tyr-701. STAT1 transcriptional regulation appeared to require activation of the canonical NF-κB pathway. Conversely, activation of the MAPK and PI3K-AKT pathways primarily regulated Ser-727 phosphorylation, whereas a physical interaction between RET/PTC3 and STAT1, followed by a direct tyrosine phosphorylation event, was necessary for STAT1 Tyr-701 phosphorylation. These data provide the first evidence of a direct link between IDO1 expression and the oncogenic activation of RET in thyroid carcinoma and describe the involved signal transduction pathways. Moreover, they suggest possible novel molecular targets for the abrogation of tumor microenvironment immunosuppression. The detection of those targets is becoming increasingly important to yield the full function of novel immune checkpoint inhibitors.
Keywords: cancer; cell signaling; indoleamine-pyrrole 2,3-dioxygenase (IDO1); interferon regulatory factor (IRF); signal transducers and activators of transcription 1 (STAT1).
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures


Similar articles
-
[Application of 9-gene panel in assisting fine needle aspiration cytology to diagnose thyroid cancer].Zhonghua Zhong Liu Za Zhi. 2024 Nov 23;46(11):1049-1057. doi: 10.3760/cma.j.cn112152-20240225-00084. Zhonghua Zhong Liu Za Zhi. 2024. PMID: 39622737 Chinese.
-
[Dose-Response Dependences for Frequency of RET/PTC Gene Rearrangements in Papillary Thyroid Carcinoma after Irradiation. Simple Pooling Analysis of Molecular Epidemiological Data].Radiats Biol Radioecol. 2016 Jan-Feb;56(1):5-25. Radiats Biol Radioecol. 2016. PMID: 27245001 Russian.
-
Tryptophan Metabolism in Obesity: The Indoleamine 2,3-Dioxygenase-1 Activity and Therapeutic Options.Adv Exp Med Biol. 2024;1460:629-655. doi: 10.1007/978-3-031-63657-8_21. Adv Exp Med Biol. 2024. PMID: 39287867 Review.
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
Defining the optimum strategy for identifying adults and children with coeliac disease: systematic review and economic modelling.Health Technol Assess. 2022 Oct;26(44):1-310. doi: 10.3310/ZUCE8371. Health Technol Assess. 2022. PMID: 36321689 Free PMC article.
Cited by
-
GD2 redirected CAR T and activated NK-cell-mediated secretion of IFNγ overcomes MYCN-dependent IDO1 inhibition, contributing to neuroblastoma cell immune escape.J Immunother Cancer. 2021 Mar;9(3):e001502. doi: 10.1136/jitc-2020-001502. J Immunother Cancer. 2021. PMID: 33737337 Free PMC article.
-
The Role of Indoleamine 2, 3-Dioxygenase 1 in Regulating Tumor Microenvironment.Cancers (Basel). 2022 Jun 1;14(11):2756. doi: 10.3390/cancers14112756. Cancers (Basel). 2022. PMID: 35681736 Free PMC article. Review.
-
Comprehensive Immune Profiling of Medullary Thyroid Cancer.Thyroid. 2020 Sep;30(9):1263-1279. doi: 10.1089/thy.2019.0604. Epub 2020 May 12. Thyroid. 2020. PMID: 32242507 Free PMC article.
-
The Immune Landscape of Thyroid Cancer in the Context of Immune Checkpoint Inhibition.Int J Mol Sci. 2019 Aug 13;20(16):3934. doi: 10.3390/ijms20163934. Int J Mol Sci. 2019. PMID: 31412566 Free PMC article. Review.
-
AXL Is a Novel Predictive Factor and Therapeutic Target for Radioactive Iodine Refractory Thyroid Cancer.Cancers (Basel). 2019 Jun 7;11(6):785. doi: 10.3390/cancers11060785. Cancers (Basel). 2019. PMID: 31181609 Free PMC article.
References
-
- Hanahan D., and Weinberg R. A. (2011) Hallmarks of cancer: the next generation. Cell 144, 646–674 - PubMed
-
- Uyttenhove C., Pilotte L., Théate I., Stroobant V., Colau D., Parmentier N., Boon T., and Van den Eynde B. J. (2003) Evidence for a tumoral immune resistance mechanism based on tryptophan degradation by indoleamine 2,3-dioxygenase. Nat. Med. 9, 1269–1274 - PubMed
-
- Mellor A. L., and Munn D. H. (2004) IDO expression by dendritic cells: tolerance and tryptophan catabolism. Nat. Rev. Immunol. 4, 762–774 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

