The enhancer binding protein Nla6 regulates developmental genes that are important for Myxococcus xanthus sporulation
- PMID: 25645554
- PMCID: PMC4352671
- DOI: 10.1128/JB.02408-14
The enhancer binding protein Nla6 regulates developmental genes that are important for Myxococcus xanthus sporulation
Abstract
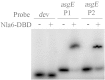
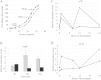
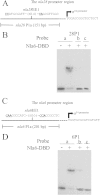
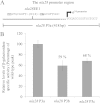
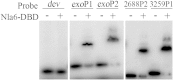
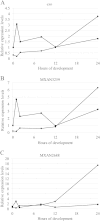
In the bacterium Myxococcus xanthus, starvation triggers the formation of multicellular fruiting bodies containing thousands of stress-resistant spores. Recent work showed that fruiting body development is regulated by a cascade of transcriptional activators called enhancer binding proteins (EBPs). The EBP Nla6 is a key component of this cascade; it regulates the promoters of other EBP genes, including a downstream-functioning EBP gene that is crucial for sporulation. In recent expression studies, hundreds of Nla6-dependent genes were identified, suggesting that the EBP gene targets of Nla6 may be part of a much larger regulon. The goal of this study was to identify and characterize genes that belong to the Nla6 regulon. Accordingly, a direct repeat [consensus, C(C/A)ACGNNGNC] binding site for Nla6 was identified using in vitro and in vivo mutational analyses, and the sequence was subsequently used to find 40 potential developmental promoter (88 gene) targets. We showed that Nla6 binds to the promoter region of four new targets (asgE, exo, MXAN2688, and MXAN3259) in vitro and that Nla6 is important for their normal expression in vivo. Phenotypic studies indicate that all of the experimentally confirmed targets of Nla6 are primarily involved in sporulation. These targets include genes involved in transcriptional regulation, cell-cell signal production, and spore differentiation and maturation. Although sporulation occurs late in development, all of the developmental loci analyzed here show an Nla6-dependent burst in expression soon after starvation is induced. This finding suggests that Nla6 starts preparing cells for sporulation very early in the developmental process.
Importance: Bacterial development yields a remarkable array of complex multicellular forms. One such form, which is commonly found in nature, is a surface-associated aggregate of cells known as a biofilm. Mature biofilms are structurally complex and contain cells that are highly resistant to antibacterial agents. When starving, the model bacterium Myxococcus xanthus forms a biofilm containing a thin mat of cells and multicellular structures that house a highly resistant cell type called a myxospore. Here, we identify the promoter binding site of the transcriptional activator Nla6, identify genes in the Nla6 regulon, and show that several of the genes in the Nla6 regulon are important for production of stress-resistant spores in starvation-induced M. xanthus biofilms.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures
Similar articles
-
Two-Component Signal Transduction Systems That Regulate the Temporal and Spatial Expression of Myxococcus xanthus Sporulation Genes.J Bacteriol. 2015 Sep 14;198(3):377-85. doi: 10.1128/JB.00474-15. Print 2016 Feb 1. J Bacteriol. 2015. PMID: 26369581 Free PMC article. Review.
-
Enhancer-binding proteins with a forkhead-associated domain and the sigma54 regulon in Myxococcus xanthus fruiting body development.Proc Natl Acad Sci U S A. 2005 Feb 22;102(8):3010-5. doi: 10.1073/pnas.0409371102. Epub 2005 Jan 24. Proc Natl Acad Sci U S A. 2005. PMID: 15668379 Free PMC article.
-
Transcription factor MrpC binds to promoter regions of hundreds of developmentally-regulated genes in Myxococcus xanthus.BMC Genomics. 2014 Dec 16;15:1123. doi: 10.1186/1471-2164-15-1123. BMC Genomics. 2014. PMID: 25515642 Free PMC article.
-
devI is an evolutionarily young negative regulator of Myxococcus xanthus development.J Bacteriol. 2015 Apr;197(7):1249-62. doi: 10.1128/JB.02542-14. Epub 2015 Feb 2. J Bacteriol. 2015. PMID: 25645563 Free PMC article.
-
Dual regulation with Ser/Thr kinase cascade and a His/Asp TCS in Myxococcus xanthus.Adv Exp Med Biol. 2008;631:111-21. doi: 10.1007/978-0-387-78885-2_7. Adv Exp Med Biol. 2008. PMID: 18792684 Review.
Cited by
-
Two-Component Signal Transduction Systems That Regulate the Temporal and Spatial Expression of Myxococcus xanthus Sporulation Genes.J Bacteriol. 2015 Sep 14;198(3):377-85. doi: 10.1128/JB.00474-15. Print 2016 Feb 1. J Bacteriol. 2015. PMID: 26369581 Free PMC article. Review.
-
Suppressor mutations reveal an NtrC-like response regulator, NmpR, for modulation of Type-IV Pili-dependent motility in Myxococcus xanthus.PLoS Genet. 2018 Oct 22;14(10):e1007714. doi: 10.1371/journal.pgen.1007714. eCollection 2018 Oct. PLoS Genet. 2018. PMID: 30346960 Free PMC article.
-
Bacterial Surface Spreading Is More Efficient on Nematically Aligned Polysaccharide Substrates.J Bacteriol. 2018 Mar 12;200(7):e00610-17. doi: 10.1128/JB.00610-17. Print 2018 Apr 1. J Bacteriol. 2018. PMID: 29311278 Free PMC article.
-
Interplay of mesoscale physics and agent-like behaviors in the parallel evolution of aggregative multicellularity.Evodevo. 2020 Oct 12;11:21. doi: 10.1186/s13227-020-00165-8. eCollection 2020. Evodevo. 2020. PMID: 33062243 Free PMC article. Review.
-
Highly Signal-Responsive Gene Regulatory Network Governing Myxococcus Development.Trends Genet. 2017 Jan;33(1):3-15. doi: 10.1016/j.tig.2016.10.006. Epub 2016 Dec 2. Trends Genet. 2017. PMID: 27916428 Free PMC article. Review.
References
-
- Kolter R. 2010. Biofilms in lab and nature: a molecular geneticist's voyage to microbial ecology. Int Microbiol 13:1–7. - PubMed
-
- Rosenberg E, Varon M. 1984. Antibiotics and lytic enzymes, p 109–125 In Rosenberg E. (ed), Myxobacteria. Development and cell interactions Springer-Verlag, New York, NY.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

