Locus-specific targeting to the X chromosome revealed by the RNA interactome of CTCF
- PMID: 25578877
- PMCID: PMC4316200
- DOI: 10.1016/j.molcel.2014.12.006
Locus-specific targeting to the X chromosome revealed by the RNA interactome of CTCF
Abstract
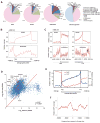
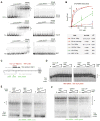
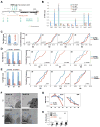
CTCF is a master regulator that plays important roles in genome architecture and gene expression. How CTCF is recruited in a locus-specific manner is not fully understood. Evidence from epigenetic processes, such as X chromosome inactivation (XCI), indicates that CTCF associates functionally with RNA. Using genome-wide approaches to investigate the relationship between its RNA interactome and epigenomic landscape, here we report that CTCF binds thousands of transcripts in mouse embryonic stem cells, many in close proximity to CTCF's genomic binding sites. CTCF is a specific and high-affinity RNA-binding protein (Kd < 1 nM). During XCI, CTCF differentially binds the active and inactive X chromosomes and interacts directly with Tsix, Xite, and Xist RNAs. Tsix and Xite RNAs target CTCF to the X inactivation center, thereby inducing homologous X chromosome pairing. Our work elucidates one mechanism by which CTCF is recruited in a locus-specific manner and implicates CTCF-RNA interactions in long-range chromosomal interactions.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures




Similar articles
-
The pluripotency factor Oct4 interacts with Ctcf and also controls X-chromosome pairing and counting.Nature. 2009 Jul 2;460(7251):128-32. doi: 10.1038/nature08098. Epub 2009 Jun 17. Nature. 2009. PMID: 19536159 Free PMC article.
-
Identification of a Ctcf cofactor, Yy1, for the X chromosome binary switch.Mol Cell. 2007 Jan 12;25(1):43-56. doi: 10.1016/j.molcel.2006.11.017. Mol Cell. 2007. PMID: 17218270
-
Evidence that homologous X-chromosome pairing requires transcription and Ctcf protein.Nat Genet. 2007 Nov;39(11):1390-6. doi: 10.1038/ng.2007.5. Epub 2007 Oct 21. Nat Genet. 2007. PMID: 17952071
-
Multifaceted role of CTCF in X-chromosome inactivation.Chromosoma. 2024 Oct;133(4):217-231. doi: 10.1007/s00412-024-00826-w. Epub 2024 Oct 21. Chromosoma. 2024. PMID: 39433641 Review.
-
CTCF: the protein, the binding partners, the binding sites and their chromatin loops.Philos Trans R Soc Lond B Biol Sci. 2013 May 6;368(1620):20120369. doi: 10.1098/rstb.2012.0369. Print 2013. Philos Trans R Soc Lond B Biol Sci. 2013. PMID: 23650640 Free PMC article. Review.
Cited by
-
Determinants and role of chromatin organization in acute leukemia.Leukemia. 2020 Oct;34(10):2561-2575. doi: 10.1038/s41375-020-0981-z. Epub 2020 Jul 20. Leukemia. 2020. PMID: 32690881 Free PMC article. Review.
-
Balancing cohesin eviction and retention prevents aberrant chromosomal interactions, Polycomb-mediated repression, and X-inactivation.Mol Cell. 2021 May 6;81(9):1970-1987.e9. doi: 10.1016/j.molcel.2021.02.031. Epub 2021 Mar 15. Mol Cell. 2021. PMID: 33725485 Free PMC article.
-
Escape from X inactivation varies in mouse tissues.PLoS Genet. 2015 Mar 18;11(3):e1005079. doi: 10.1371/journal.pgen.1005079. eCollection 2015 Mar. PLoS Genet. 2015. PMID: 25785854 Free PMC article.
-
Emerging Roles of Repetitive and Repeat-Containing RNA in Nuclear and Chromatin Organization and Gene Expression.Front Cell Dev Biol. 2021 Oct 6;9:735527. doi: 10.3389/fcell.2021.735527. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34722514 Free PMC article. Review.
-
Regulatory feedback from nascent RNA to chromatin and transcription.Nat Rev Mol Cell Biol. 2017 May;18(5):331-337. doi: 10.1038/nrm.2017.12. Epub 2017 Mar 8. Nat Rev Mol Cell Biol. 2017. PMID: 28270684 Review.
References
-
- Bacher CP, Guggiari M, Brors B, Augui S, Clerc P, Avner P, Eils R, Heard E. Transient colocalization of X-inactivation centres accompanies the initiation of X inactivation. Nat Cell Biol. 2006;8:293–299. - PubMed
-
- Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

