Mutation of the melastatin-related cation channel, TRPM3, underlies inherited cataract and glaucoma
- PMID: 25090642
- PMCID: PMC4121231
- DOI: 10.1371/journal.pone.0104000
Mutation of the melastatin-related cation channel, TRPM3, underlies inherited cataract and glaucoma
Abstract
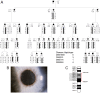
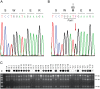
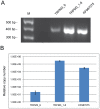
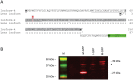
Inherited forms of cataract are a clinically important and genetically heterogeneous cause of visual impairment that usually present at an early age with or without systemic and/or other ocular abnormalities. Here we have identified a new locus for inherited cataract and high-tension glaucoma with variable anterior segment defects, and characterized an underlying mutation in the gene coding for transient receptor potential cation channel, subfamily M, member-3 (TRPM3, melastatin-2). Genome-wide linkage analysis mapped the ocular disease locus to the pericentric region of human chromosome 9. Whole exome and custom-target next-generation sequencing detected a heterozygous A-to-G transition in exon-3 of TRPM3 that co-segregated with disease. As a consequence of alternative splicing this missense mutation was predicted to result in the substitution of isoleucine-to-methionine at codon 65 (c.195A>G; p.I65 M) of TRPM3 transcript variant 9, and at codon 8 (c.24A>G; p.I8 M) of a novel TRPM3 transcript variant expressed in human lens. In both transcript variants the I-to-M substitution was predicted in silico to exert damaging effects on protein function. Furthermore, transient expression studies of a recombinant TRPM3-GFP reporter product predicted that the I-to-M substitution introduced an alternative translation start-site located 89 codons upstream from the native initiator methionine found in eight other TRPM3 transcript variants (1-8). Collectively, these studies have provided the first evidence that TRPM3 is associated with inherited ocular disease in humans, and further provide support for the important role of this cation channel in normal eye development.
Conflict of interest statement
Figures

Similar articles
-
Mutation of the TRPM3 cation channel underlies progressive cataract development and lens calcification associated with pro-fibrotic and immune cell responses.FASEB J. 2021 Feb;35(2):e21288. doi: 10.1096/fj.202002037R. FASEB J. 2021. PMID: 33484482 Free PMC article.
-
A novel beaded filament structural protein 1 (BFSP1) gene mutation associated with autosomal dominant congenital cataract in a Chinese family.Mol Vis. 2013 Dec 27;19:2590-5. eCollection 2013. Mol Vis. 2013. PMID: 24379646 Free PMC article.
-
Cerulean cataract mapped to 12q13 and associated with a novel initiation codon mutation in MIP.Mol Vis. 2011;17:2049-55. Epub 2011 Jul 26. Mol Vis. 2011. PMID: 21850180 Free PMC article.
-
TRPM3_miR-204: a complex locus for eye development and disease.Hum Genomics. 2020 Feb 18;14(1):7. doi: 10.1186/s40246-020-00258-4. Hum Genomics. 2020. PMID: 32070426 Free PMC article. Review.
-
TRPM3 in the eye and in the nervous system - from new findings to novel mechanisms.Biol Chem. 2022 Mar 2;403(8-9):859-868. doi: 10.1515/hsz-2021-0403. Print 2022 Jul 26. Biol Chem. 2022. PMID: 35240732 Review.
Cited by
-
TRPM3 in temperature sensing and beyond.Temperature (Austin). 2015 Feb 25;2(2):201-13. doi: 10.4161/23328940.2014.988524. eCollection 2015 Apr-Jun. Temperature (Austin). 2015. PMID: 27227024 Free PMC article. Review.
-
Functions of TRPs in retinal tissue in physiological and pathological conditions.Front Mol Neurosci. 2024 Sep 25;17:1459083. doi: 10.3389/fnmol.2024.1459083. eCollection 2024. Front Mol Neurosci. 2024. PMID: 39386050 Free PMC article. Review.
-
Connexin Mutants Compromise the Lens Circulation and Cause Cataracts through Biomineralization.Int J Mol Sci. 2020 Aug 13;21(16):5822. doi: 10.3390/ijms21165822. Int J Mol Sci. 2020. PMID: 32823750 Free PMC article. Review.
-
Through the Cat-Map Gateway: A Brief History of Cataract Genetics.Genes (Basel). 2024 Jun 14;15(6):785. doi: 10.3390/genes15060785. Genes (Basel). 2024. PMID: 38927721 Free PMC article. Review.
-
Inherited cataracts: Genetic mechanisms and pathways new and old.Exp Eye Res. 2021 Aug;209:108662. doi: 10.1016/j.exer.2021.108662. Epub 2021 Jun 12. Exp Eye Res. 2021. PMID: 34126080 Free PMC article. Review.
References
-
- Trumler AA (2011) Evaluation of pediatric cataracts and systemic disorders. Curr Opin Ophthalmol 22: 365–379. - PubMed
-
- Rahi JS, Dezateux C (2001) Measuring and interpreting the incidence of congenital ocular anomalies: lessons from a national study of congenital cataract in the UK. Invest Ophthalmol Vis Sci 42: 1444–1448. - PubMed
-
- Amaya L, Taylor D, Russell-Eggitt I, Nischal KK, Lengyel D (2003) The morphology and natural history of childhood cataracts. Surv Ophthalmol 48: 125–44. - PubMed
-
- Zetterstrom C, Lundvall A, Kugelberg M (2005) Cataracts in children. J Cataract Refract Surg 31: 824–840. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

