Delimiting cryptic pathogen species causing apple Valsa canker with multilocus data
- PMID: 24834333
- PMCID: PMC4020696
- DOI: 10.1002/ece3.1030
Delimiting cryptic pathogen species causing apple Valsa canker with multilocus data
Abstract
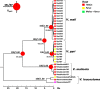
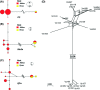
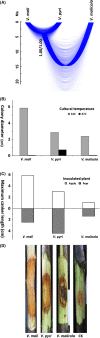
Fungal diseases are posing tremendous threats to global economy and food safety. Among them, Valsa canker, caused by fungi of Valsa and their Cytospora anamorphs, has been a serious threat to fruit and forest trees and is one of the most destructive diseases of apple in East Asia, particularly. Accurate and robust delimitation of pathogen species is not only essential for the development of effective disease control programs, but also will advance our understanding of the emergence of plant diseases. However, species delimitation is especially difficult in Valsa because of the high variability of morphological traits and in many cases the lack of the teleomorph. In this study, we delimitated species boundary for pathogens causing apple Valsa canker with a multifaceted approach. Based on three independent loci, the internal transcribed spacer (ITS), β-tubulin (Btu), and translation elongation factor-1 alpha (EF1α), we inferred gene trees with both maximum likelihood and Bayesian methods, estimated species tree with Bayesian multispecies coalescent approaches, and validated species tree with Bayesian species delimitation. Through divergence time estimation and ancestral host reconstruction, we tested the possible underlying mechanisms for fungal speciation and host-range change. Our results proved that two varieties of the former morphological species V. mali represented two distinct species, V. mali and V. pyri, which diverged about 5 million years ago, much later than the divergence of their preferred hosts, excluding a scenario of fungi-host co-speciation. The marked different thermal preferences and contrasting pathogenicity in cross-inoculation suggest ecological divergences between the two species. Apple was the most likely ancestral host for both V. mali and V. pyri. Host-range expansion led to the occurrence of V. pyri on both pear and apple. Our results also represent an example in which ITS data might underestimate species diversity.
Keywords: Valsa; ancestral host reconstruction; host-range expansion; molecular dating; species delimitation.
Figures

Similar articles
-
Re-evaluation of pathogens causing Valsa canker on apple in China.Mycologia. 2011 Mar-Apr;103(2):317-24. doi: 10.3852/09-165. Epub 2010 Oct 7. Mycologia. 2011. PMID: 21415290
-
First Report of Valsa leucostoma Causing Valsa Canker of Pyrus communis (cv. Duchess de' Angouleme) in China.Plant Dis. 2014 Mar;98(3):422. doi: 10.1094/PDIS-07-13-0704-PDN. Plant Dis. 2014. PMID: 30708418
-
A Nested PCR Assay for Detecting Valsa mali var. mali in Different Tissues of Apple Trees.Plant Dis. 2012 Nov;96(11):1645-1652. doi: 10.1094/PDIS-05-11-0387-RE. Plant Dis. 2012. PMID: 30727457
-
First Report of Valsa nivea (Hoffm.) Fr. Causing Valsa Canker on Korla Pear (Pyrus sinkiangensis Yü) in the World.Plant Dis. 2023 Feb 23. doi: 10.1094/PDIS-09-22-2038-PDN. Online ahead of print. Plant Dis. 2023. PMID: 36825310
-
Identification of Fungal Species Associated with Apple Canker in Tarim Basin, China.Plant Dis. 2023 May;107(5):1284-1298. doi: 10.1094/PDIS-09-22-2032-SR. Epub 2023 May 8. Plant Dis. 2023. PMID: 36281021
Cited by
-
Apple crown and collar canker and necrosis caused by Cytospora balanejica sp. nov. in Iran.Sci Rep. 2024 Mar 19;14(1):6629. doi: 10.1038/s41598-024-57235-3. Sci Rep. 2024. PMID: 38504125 Free PMC article.
-
Two members of the velvet family, VmVeA and VmVelB, affect conidiation, virulence and pectinase expression in Valsa mali.Mol Plant Pathol. 2018 Jul;19(7):1639-1651. doi: 10.1111/mpp.12645. Epub 2018 Feb 13. Mol Plant Pathol. 2018. PMID: 29127722 Free PMC article.
-
Agrilus mali Matsumara (Coleoptera: Buprestidae), a new invasive pest of wild apple in western China: DNA barcoding and life cycle.Ecol Evol. 2018 Dec 27;9(3):1160-1172. doi: 10.1002/ece3.4804. eCollection 2019 Feb. Ecol Evol. 2018. PMID: 30805149 Free PMC article.
-
Vm-milR37 contributes to pathogenicity by regulating glutathione peroxidase gene VmGP in Valsa mali.Mol Plant Pathol. 2021 Feb;22(2):243-254. doi: 10.1111/mpp.13023. Epub 2020 Dec 5. Mol Plant Pathol. 2021. PMID: 33278058 Free PMC article.
-
A Novel DCL2-Dependent Micro-Like RNA Vm-PC-3p-92107_6 Affects Pathogenicity by Regulating the Expression of Vm-VPS10 in Valsa mali.Front Microbiol. 2021 Oct 1;12:721399. doi: 10.3389/fmicb.2021.721399. eCollection 2021. Front Microbiol. 2021. PMID: 34759897 Free PMC article.
References
-
- Abe K, Kotoda N, Kato H, Soejima J. Resistance sources to Valsa canker (Valsa ceratosperma) in a germplasm collection of diverse Malus species. Plant Breeding. 2007;126:449–453.
-
- Adams GC, Surve-Iyer RS, Iezzoni A. Ribosomal DNA sequence divergence and group I introns within the Leucostoma species L. cinctum L. persoonii and L. parapersoonii sp. nov., ascomycetes that cause Cytospora canker of fruit trees. Mycologia. 2002;94(6):947–967. - PubMed
-
- Agrios GN. Plant Pathology. San Diego, CA: Academic Press; 1997.
-
- Bandelt HJ, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999;16:37–48. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

