Two DHH subfamily 1 proteins in Streptococcus pneumoniae possess cyclic di-AMP phosphodiesterase activity and affect bacterial growth and virulence
- PMID: 24013631
- PMCID: PMC3811582
- DOI: 10.1128/JB.00769-13
Two DHH subfamily 1 proteins in Streptococcus pneumoniae possess cyclic di-AMP phosphodiesterase activity and affect bacterial growth and virulence
Abstract
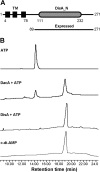
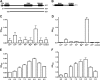
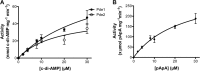
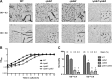
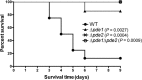
Cyclic di-AMP (c-di-AMP) and cyclic di-GMP (c-di-GMP) are signaling molecules that play important roles in bacterial biology and pathogenesis. However, these nucleotides have not been explored in Streptococcus pneumoniae, an important bacterial pathogen. In this study, we characterized the c-di-AMP-associated genes of S. pneumoniae. The results showed that SPD_1392 (DacA) is a diadenylate cyclase that converts ATP to c-di-AMP. Both SPD_2032 (Pde1) and SPD_1153 (Pde2), which belong to the DHH subfamily 1 proteins, displayed c-di-AMP phosphodiesterase activity. Pde1 cleaved c-di-AMP into phosphoadenylyl adenosine (pApA), whereas Pde2 directly hydrolyzed c-di-AMP into AMP. Additionally, Pde2, but not Pde1, degraded pApA into AMP. Our results also demonstrated that both Pde1 and Pde2 played roles in bacterial growth, resistance to UV treatment, and virulence in a mouse pneumonia model. These results indicate that c-di-AMP homeostasis is essential for pneumococcal biology and disease.
Figures


Similar articles
-
Cyclic di-AMP impairs potassium uptake mediated by a cyclic di-AMP binding protein in Streptococcus pneumoniae.J Bacteriol. 2014 Feb;196(3):614-23. doi: 10.1128/JB.01041-13. Epub 2013 Nov 22. J Bacteriol. 2014. PMID: 24272783 Free PMC article.
-
Bacterial Second Messenger Cyclic di-AMP Modulates the Competence State in Streptococcus pneumoniae.J Bacteriol. 2020 Jan 29;202(4):e00691-19. doi: 10.1128/JB.00691-19. Print 2020 Jan 29. J Bacteriol. 2020. PMID: 31767779 Free PMC article.
-
Unique Roles for Streptococcus pneumoniae Phosphodiesterase 2 in Cyclic di-AMP Catabolism and Macrophage Responses.Front Immunol. 2020 Mar 31;11:554. doi: 10.3389/fimmu.2020.00554. eCollection 2020. Front Immunol. 2020. PMID: 32300347 Free PMC article.
-
Bacterial second messenger cyclic di-AMP in streptococci.Mol Microbiol. 2023 Dec;120(6):791-804. doi: 10.1111/mmi.15187. Epub 2023 Oct 28. Mol Microbiol. 2023. PMID: 37898560 Review.
-
Making and Breaking of an Essential Poison: the Cyclases and Phosphodiesterases That Produce and Degrade the Essential Second Messenger Cyclic di-AMP in Bacteria.J Bacteriol. 2018 Dec 7;201(1):e00462-18. doi: 10.1128/JB.00462-18. Print 2019 Jan 1. J Bacteriol. 2018. PMID: 30224435 Free PMC article. Review.
Cited by
-
The role of bacterial cyclic di-adenosine monophosphate in the host immune response.Front Microbiol. 2022 Aug 29;13:958133. doi: 10.3389/fmicb.2022.958133. eCollection 2022. Front Microbiol. 2022. PMID: 36106081 Free PMC article. Review.
-
The Second Messenger c-di-AMP Regulates Diverse Cellular Pathways Involved in Stress Response, Biofilm Formation, Cell Wall Homeostasis, SpeB Expression, and Virulence in Streptococcus pyogenes.Infect Immun. 2019 May 21;87(6):e00147-19. doi: 10.1128/IAI.00147-19. Print 2019 Jun. Infect Immun. 2019. PMID: 30936159 Free PMC article.
-
c-di-AMP hydrolysis by the phosphodiesterase AtaC promotes differentiation of multicellular bacteria.Proc Natl Acad Sci U S A. 2020 Mar 31;117(13):7392-7400. doi: 10.1073/pnas.1917080117. Epub 2020 Mar 18. Proc Natl Acad Sci U S A. 2020. PMID: 32188788 Free PMC article.
-
The c-di-AMP signaling system influences stress tolerance and biofilm formation of Streptococcus mitis.Microbiologyopen. 2021 Aug;10(4):e1203. doi: 10.1002/mbo3.1203. Microbiologyopen. 2021. PMID: 34459556 Free PMC article.
-
New Insights into the Cyclic Di-adenosine Monophosphate (c-di-AMP) Degradation Pathway and the Requirement of the Cyclic Dinucleotide for Acid Stress Resistance in Staphylococcus aureus.J Biol Chem. 2016 Dec 30;291(53):26970-26986. doi: 10.1074/jbc.M116.747709. Epub 2016 Nov 10. J Biol Chem. 2016. PMID: 27834680 Free PMC article.
References
-
- Hengge R. 2009. Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 7:263–273 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

