CDK8-mediated STAT1-S727 phosphorylation restrains NK cell cytotoxicity and tumor surveillance
- PMID: 23933255
- PMCID: PMC3748339
- DOI: 10.1016/j.celrep.2013.07.012
CDK8-mediated STAT1-S727 phosphorylation restrains NK cell cytotoxicity and tumor surveillance
Abstract
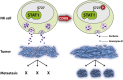
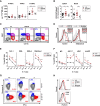
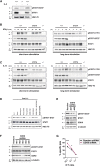
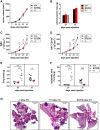
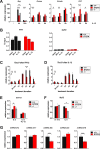
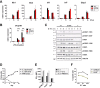
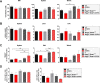
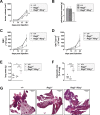
The transcription factor STAT1 is important in natural killer (NK) cells, which provide immediate defense against tumor and virally infected cells. We show that mutation of a single phosphorylation site (Stat1-S727A) enhances NK cell cytotoxicity against a range of tumor cells, accompanied by increased expression of perforin and granzyme B. Stat1-S727A mice display significantly delayed disease onset in NK cell-surveilled tumor models including melanoma, leukemia, and metastasizing breast cancer. Constitutive phosphorylation of S727 depends on cyclin-dependent kinase 8 (CDK8). Inhibition of CDK8-mediated STAT1-S727 phosphorylation may thus represent a therapeutic strategy for stimulating NK cell-mediated tumor surveillance.
Copyright © 2013 The Authors. Published by Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Selective and Potent CDK8/19 Inhibitors Enhance NK-Cell Activity and Promote Tumor Surveillance.Mol Cancer Ther. 2020 Apr;19(4):1018-1030. doi: 10.1158/1535-7163.MCT-19-0789. Epub 2020 Feb 5. Mol Cancer Ther. 2020. PMID: 32024684 Free PMC article.
-
NK Cell-Specific CDK8 Deletion Enhances Antitumor Responses.Cancer Immunol Res. 2018 Apr;6(4):458-466. doi: 10.1158/2326-6066.CIR-17-0183. Epub 2018 Jan 31. Cancer Immunol Res. 2018. PMID: 29386186
-
CDK8 kinase phosphorylates transcription factor STAT1 to selectively regulate the interferon response.Immunity. 2013 Feb 21;38(2):250-62. doi: 10.1016/j.immuni.2012.10.017. Epub 2013 Jan 24. Immunity. 2013. PMID: 23352233 Free PMC article.
-
STAT1 Isoforms Differentially Regulate NK Cell Maturation and Anti-tumor Activity.Front Immunol. 2020 Sep 11;11:2189. doi: 10.3389/fimmu.2020.02189. eCollection 2020. Front Immunol. 2020. PMID: 33042133 Free PMC article.
-
STAT1-S727 - the license to kill.Oncoimmunology. 2014 Dec 13;3(9):e955441. doi: 10.4161/21624011.2014.955441. eCollection 2014 Oct. Oncoimmunology. 2014. PMID: 25941617 Free PMC article.
Cited by
-
Interferon Independent Non-Canonical STAT Activation and Virus Induced Inflammation.Viruses. 2018 Apr 14;10(4):196. doi: 10.3390/v10040196. Viruses. 2018. PMID: 29662014 Free PMC article. Review.
-
p35 is a Crucial Player in NK-cell Cytotoxicity and TGFβ-mediated NK-cell Dysfunction.Cancer Res Commun. 2023 May 5;3(5):793-806. doi: 10.1158/2767-9764.CRC-22-0497. eCollection 2023 May. Cancer Res Commun. 2023. PMID: 37377891 Free PMC article.
-
Response to interferons and antibacterial innate immunity in the absence of tyrosine-phosphorylated STAT1.EMBO Rep. 2016 Mar;17(3):367-82. doi: 10.15252/embr.201540726. Epub 2016 Feb 12. EMBO Rep. 2016. PMID: 26882544 Free PMC article.
-
Regulation of metabolism by the Mediator complex.Biophys Rep. 2016;2(2):69-77. doi: 10.1007/s41048-016-0031-6. Epub 2016 Nov 1. Biophys Rep. 2016. PMID: 28018965 Free PMC article.
-
TNFR2/BIRC3-TRAF1 signaling pathway as a novel NK cell immune checkpoint in cancer.Oncoimmunology. 2017 Oct 11;7(12):e1386826. doi: 10.1080/2162402X.2017.1386826. eCollection 2018. Oncoimmunology. 2017. PMID: 30524877 Free PMC article.
References
-
- Cao X., Shores E.W., Hu-Li J., Anver M.R., Kelsall B.L., Russell S.M., Drago J., Noguchi M., Grinberg A., Bloom E.T. Defective lymphoid development in mice lacking expression of the common cytokine receptor gamma chain. Immunity. 1995;2:223–238. - PubMed
-
- Dalton D.K., Pitts-Meek S., Keshav S., Figari I.S., Bradley A., Stewart T.A. Multiple defects of immune cell function in mice with disrupted interferon-gamma genes. Science. 1993;259:1739–1742. - PubMed
-
- Davis M.I., Hunt J.P., Herrgard S., Ciceri P., Wodicka L.M., Pallares G., Hocker M., Treiber D.K., Zarrinkar P.P. Comprehensive analysis of kinase inhibitor selectivity. Nat. Biotechnol. 2011;29:1046–1051. - PubMed
Supplemental References
-
- Gratz N., Siller M., Schaljo B., Pirzada Z.A., Gattermeier I., Vojtek I., Kirschning C.J., Wagner H., Akira S., Charpentier E., Kovarik P. Group A streptococcus activates type I interferon production and MyD88-dependent signaling without involvement of TLR2, TLR4, and TLR9. J. Biol. Chem. 2008;283:19879–19887. - PMC - PubMed
-
- Kamezaki K., Shimoda K., Numata A., Matsuda T., Nakayama K.I., Harada M. The role of Tyk2, Stat1 and Stat4 in LPS-induced endotoxin signals. Int. Immunol. 2004;16:1173–1179. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

