Somatic SETBP1 mutations in myeloid malignancies
- PMID: 23832012
- PMCID: PMC3729750
- DOI: 10.1038/ng.2696
Somatic SETBP1 mutations in myeloid malignancies
Abstract
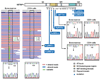
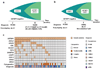
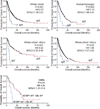
Here we report whole-exome sequencing of individuals with various myeloid malignancies and identify recurrent somatic mutations in SETBP1, consistent with a recent report on atypical chronic myeloid leukemia (aCML). Closely positioned somatic SETBP1 mutations encoding changes in Asp868, Ser869, Gly870, Ile871 and Asp880, which match germline mutations in Schinzel-Giedion syndrome (SGS), were detected in 17% of secondary acute myeloid leukemias (sAML) and 15% of chronic myelomonocytic leukemia (CMML) cases. These results from deep sequencing demonstrate a higher mutational detection rate than reported with conventional sequencing methodology. Mutant cases were associated with advanced age and monosomy 7/deletion 7q (-7/del(7q)) constituting poor prognostic factors. Analysis of serially collected samples indicated that SETBP1 mutations were acquired during leukemic evolution. Transduction with mutant Setbp1 led to the immortalization of mouse myeloid progenitors that showed enhanced proliferative capacity compared to cells transduced with wild-type Setbp1. Somatic mutations of SETBP1 seem to cause gain of function, are associated with myeloid leukemic transformation and convey poor prognosis in myelodysplastic syndromes (MDS) and CMML.
Figures



Comment in
-
A new player SETs in myeloid malignancy.Nat Genet. 2013 Aug;45(8):846-7. doi: 10.1038/ng.2709. Nat Genet. 2013. PMID: 23892662 Free PMC article.
Similar articles
-
Somatic SETBP1 mutations in myeloid neoplasms.Int J Hematol. 2017 Jun;105(6):732-742. doi: 10.1007/s12185-017-2241-1. Epub 2017 Apr 26. Int J Hematol. 2017. PMID: 28447248 Review.
-
Mutations in SETBP1 are recurrent in myelodysplastic syndromes and often coexist with cytogenetic markers associated with disease progression.Br J Haematol. 2013 Oct;163(2):235-9. doi: 10.1111/bjh.12491. Epub 2013 Jul 24. Br J Haematol. 2013. PMID: 23889083
-
Only SETBP1 hotspot mutations are associated with refractory disease in myeloid malignancies.J Cancer Res Clin Oncol. 2017 Dec;143(12):2511-2519. doi: 10.1007/s00432-017-2518-z. Epub 2017 Sep 14. J Cancer Res Clin Oncol. 2017. PMID: 28913558
-
SETBP1 mutations occur in 9% of MDS/MPN and in 4% of MPN cases and are strongly associated with atypical CML, monosomy 7, isochromosome i(17)(q10), ASXL1 and CBL mutations.Leukemia. 2013 Sep;27(9):1852-60. doi: 10.1038/leu.2013.133. Epub 2013 Apr 30. Leukemia. 2013. PMID: 23628959
-
SETBP1 mutations as a biomarker for myelodysplasia /myeloproliferative neoplasm overlap syndrome.Biomark Res. 2017 Dec 6;5:33. doi: 10.1186/s40364-017-0113-8. eCollection 2017. Biomark Res. 2017. PMID: 29225884 Free PMC article. Review.
Cited by
-
An international consortium proposal of uniform response criteria for myelodysplastic/myeloproliferative neoplasms (MDS/MPN) in adults.Blood. 2015 Mar 19;125(12):1857-65. doi: 10.1182/blood-2014-10-607341. Epub 2015 Jan 26. Blood. 2015. PMID: 25624319 Free PMC article.
-
BRCC3 mutations in myeloid neoplasms.Haematologica. 2015 Aug;100(8):1051-7. doi: 10.3324/haematol.2014.111989. Epub 2015 May 22. Haematologica. 2015. PMID: 26001790 Free PMC article.
-
Multivariate Analysis Identifies Eight Novel Loci Associated with Meat Productivity Traits in Sheep.Genes (Basel). 2021 Mar 4;12(3):367. doi: 10.3390/genes12030367. Genes (Basel). 2021. PMID: 33806625 Free PMC article.
-
Genome-wide association study of antibody level response to NDV and IBV in Jinghai yellow chicken based on SLAF-seq technology.J Appl Genet. 2015 Aug;56(3):365-73. doi: 10.1007/s13353-014-0269-y. Epub 2015 Jan 15. J Appl Genet. 2015. PMID: 25588649
-
Molecular genetics and management of world health organization defined atypical chronic myeloid leukemia.Ann Hematol. 2023 Apr;102(4):777-785. doi: 10.1007/s00277-023-05106-8. Epub 2023 Feb 3. Ann Hematol. 2023. PMID: 36735076
References
-
- Hoischen A, et al. De novo mutations of SETBP1 cause Schinzel-Giedion syndrome. Nat Genet. 2010;42:483–485. - PubMed
-
- Damm F, et al. SETBP1 mutations in 658 patients with myelodysplastic syndromes, chronic myelomonocytic leukemia and secondary acute myeloid leukemias. Leukemia. 2013 - PubMed
-
- Thol F, et al. SETBP1 mutation analysis in 944 patients with MDS and AML. Leukemia. 2013 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

